BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= QCG11e12.yg.2.1
(636 letters)
Database: /db/uniprot/tmp/swall
1,395,590 sequences; 442,889,342 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
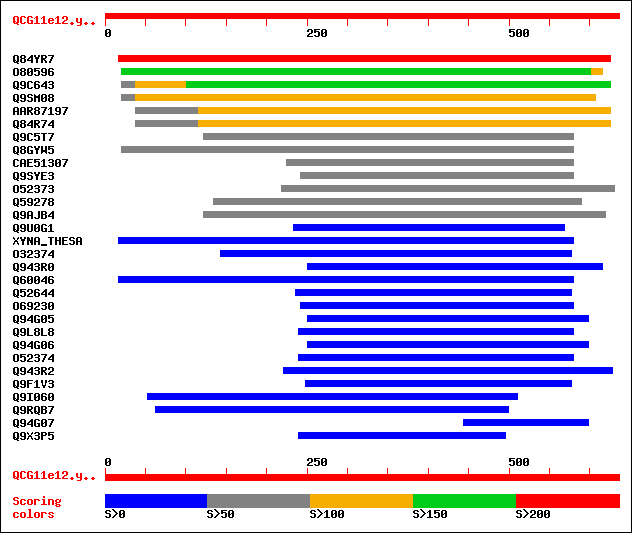
sptr|Q84YR7|Q84YR7 Putative 1,4-beta-D xylan xylanohydrolase. 322 3e-87
sptr|O80596|O80596 T27I1.7 protein. 155 5e-37
sptr|Q9C643|Q9C643 Xylan endohydrolase isoenzyme, putative. 152 5e-36
sptr|Q9SM08|Q9SM08 RXF12 protein (F19C14.2 protein) (Xylan endoh... 149 2e-35
sptrnew|AAR87197|AAR87197 Putative xylan xylanohydrolase. 101 9e-21
sptr|Q84R74|Q84R74 Putative xylan xylanohydrolase isoenzyme. 101 9e-21
sptr|Q9C5T7|Q9C5T7 Putative xylanase containing two cellulose bi... 89 5e-17
sptr|Q8GYW5|Q8GYW5 Putative xylan endohydrolase (At4g08160). 89 5e-17
sptrnew|CAE51307|CAE51307 Beta-1,4-xylanase. 72 8e-12
sptr|Q9SYE3|Q9SYE3 Putative xylan endohydrolase. 69 5e-11
sptr|O52373|O52373 Family 10 xylanase (EC 3.2.1.8). 62 6e-09
sptr|Q59278|Q59278 Endoxylanase (EC 3.2.1.8). 53 3e-06
sptr|Q9AJB4|Q9AJB4 Xylanase B (EC 3.2.1.8) (Endo-1,4-beta-xylana... 52 6e-06
sptr|Q9U0G1|Q9U0G1 Xylanase B precursor. 49 4e-05
sw|P36917|XYNA_THESA Endo-1,4-beta-xylanase A precursor (EC 3.2.... 47 2e-04
sptr|O32374|O32374 Xylanase precursor. 47 2e-04
sptr|Q943R0|Q943R0 Putative endo-1,4-beta-xylanase X-1. 47 2e-04
sptr|Q60046|Q60046 XynA precursor (EC 3.2.1.8). 47 3e-04
sptr|Q52644|Q52644 XynA (EC 3.2.1.8) (Endo-1,4-beta-xylanase). 46 4e-04
sptr|O69230|O69230 Endo-1,4-beta-xylanase (EC 3.2.1.8). 45 8e-04
sptr|Q94G05|Q94G05 1,4-beta-D xylan xylanohydrolase. 44 0.001
sptr|Q9L8L8|Q9L8L8 Beta-1,4-xylanase XynA precursor. 44 0.001
sptr|Q94G06|Q94G06 1,4-beta-D xylan xylanohydrolase. 44 0.002
sptr|O52374|O52374 Family 10 xylanase (EC 3.2.1.8). 43 0.003
sptr|Q943R2|Q943R2 Putative endo-1,4-beta-xylanase X-1. 42 0.005
sptr|Q9F1V3|Q9F1V3 Xylanase A. 42 0.009
sptr|Q9I060|Q9I060 Hypothetical protein PA2783. 41 0.011
sptr|Q9RQB7|Q9RQB7 Endo-1,4-beta-xylanase (EC 3.2.1.8). 41 0.011
sptr|Q94G07|Q94G07 1,4-beta-D xylan xylanohydrolase (Fragment). 40 0.019
sptr|Q9X3P5|Q9X3P5 XynA. 40 0.019
>sptr|Q84YR7|Q84YR7 Putative 1,4-beta-D xylan xylanohydrolase.
Length = 1101
Score = 322 bits (825), Expect = 3e-87
Identities = 159/207 (76%), Positives = 175/207 (84%), Gaps = 4/207 (1%)
Frame = +2
Query: 17 RRLXXXXEGPPPGVDLLIDSVTISYKKTASS----VGGTENIILNYDFSKGLHPWNPIRC 184
RRL EGPPPGVDLLIDSVTISYKKT + V GTENII NYDFS+GLH WNPI C
Sbjct: 180 RRLVFYIEGPPPGVDLLIDSVTISYKKTERAASKLVSGTENIISNYDFSEGLHLWNPICC 239
Query: 185 HAYAASQWSGFLDGIRGNSGENYAVVSKRTEHWQGLEQDITNQVSTGTVYVVSAFVRVDG 364
HAY ASQWSGFLDGIRG+SGENYAVVSKRTE WQGLEQDIT++VS GT Y VSA+VRVDG
Sbjct: 240 HAYVASQWSGFLDGIRGSSGENYAVVSKRTESWQGLEQDITDKVSAGTAYAVSAYVRVDG 299
Query: 365 IVQGQVEVKATLRLQNADGSTHYNPVGSVVASKEKWNKLEGSFSLTNMPKNVVFYLEGPP 544
+ +VEVKATLRL N D STHY+PVGS++ASKEKW K+EGSF LTNMPK VVFYLEGPP
Sbjct: 300 NIHTKVEVKATLRLHNTDDSTHYSPVGSLLASKEKWEKMEGSFCLTNMPKRVVFYLEGPP 359
Query: 545 AGVDLVIDSVTVACSRHKQSKEVKVPA 625
AGVDL+IDSV V CS ++Q KEVKVP+
Sbjct: 360 AGVDLIIDSVNVTCSGYQQLKEVKVPS 386
Score = 180 bits (457), Expect = 1e-44
Identities = 84/161 (52%), Positives = 118/161 (73%)
Frame = +2
Query: 122 ENIILNYDFSKGLHPWNPIRCHAYAASQWSGFLDGIRGNSGENYAVVSKRTEHWQGLEQD 301
ENI+ N DFS+GLH W+P CH + A + SG+ GIR +SG NYAV+++RT +WQGLEQD
Sbjct: 50 ENILSNNDFSEGLHLWHPNGCHGFVAVEGSGYHHGIRPHSGSNYAVLTRRTHNWQGLEQD 109
Query: 302 ITNQVSTGTVYVVSAFVRVDGIVQGQVEVKATLRLQNADGSTHYNPVGSVVASKEKWNKL 481
IT +V+ GT Y+V+A VRV G + V ++ATL+L+ ST+Y V + ASK+ W KL
Sbjct: 110 ITEKVTVGTEYIVAAHVRVHGELNEPVGIQATLKLEGDGSSTNYQSVARISASKDCWEKL 169
Query: 482 EGSFSLTNMPKNVVFYLEGPPAGVDLVIDSVTVACSRHKQS 604
EGSF L +P+ +VFY+EGPP GVDL+IDSVT++ + +++
Sbjct: 170 EGSFELKTLPRRLVFYIEGPPPGVDLLIDSVTISYKKTERA 210
Score = 101 bits (252), Expect = 7e-21
Identities = 61/197 (30%), Positives = 101/197 (51%), Gaps = 5/197 (2%)
Frame = +2
Query: 17 RRLXXXXEGPPPGVDLLIDSVTISYK-----KTASSVGGTENIILNYDFSKGLHPWNPIR 181
+R+ EGPP GVDL+IDSV ++ K G + I+ N F +GL+ W+
Sbjct: 349 KRVVFYLEGPPAGVDLIIDSVNVTCSGYQQLKEVKVPSGVDTIVKNPHFDEGLNNWSGRG 408
Query: 182 CHAYAASQWSGFLDGIRGNSGENYAVVSKRTEHWQGLEQDITNQVSTGTVYVVSAFVRVD 361
C+ + ++ +G +A + R +W G++QDIT +V +Y +S+ VR+
Sbjct: 409 CNICRHELTA--YGNVKPLNGSYFASATGRVHNWNGIQQDITGRVQRKVLYEISSAVRIF 466
Query: 362 GIVQGQVEVKATLRLQNADGSTHYNPVGSVVASKEKWNKLEGSFSLTNMPKNVVFYLEGP 541
G EV TL +Q G Y + AS ++W L+G F L V ++EGP
Sbjct: 467 GSAN-DTEVCVTLWVQEY-GRERYVSLAKNPASDKQWTHLKGKFLLHAPFSKAVIFVEGP 524
Query: 542 PAGVDLVIDSVTVACSR 592
PAG+D+++D + ++ +R
Sbjct: 525 PAGIDILVDGLVLSPAR 541
Score = 88.2 bits (217), Expect = 8e-17
Identities = 61/193 (31%), Positives = 91/193 (47%), Gaps = 19/193 (9%)
Frame = +2
Query: 38 EGPPPGVDLLIDSVTISYKKTA--------SSVGGTENIILNYDFSKGLHPWNP-----I 178
EGPP G+D+L+D + +S + +V N+I N FS GL W+P +
Sbjct: 522 EGPPAGIDILVDGLVLSPARKLHAAPRPRIENVSYGANVIHNSAFSHGLSGWSPMGSCRL 581
Query: 179 RCHAYAASQWSGFLDG------IRGNSGENYAVVSKRTEHWQGLEQDITNQVSTGTVYVV 340
H + S L IRG+ Y + + RT+ W G Q IT+++ T Y V
Sbjct: 582 SIHTESPHMLSAILKDPSAKQHIRGS----YILATNRTDVWMGPSQLITDKLRLHTTYRV 637
Query: 341 SAFVRVDGIVQGQVEVKATLRLQNADGSTHYNPVGSVVASKEKWNKLEGSFSLTNMPKNV 520
SA+VR G+ V L + + + G V A ++W +L+G+F L P V
Sbjct: 638 SAWVRAGSGGHGRYHVNVCLAVDH-----QWVNGGQVEADGDQWYELKGAFKLEKKPSKV 692
Query: 521 VFYLEGPPAGVDL 559
Y++GPP GVDL
Sbjct: 693 TAYVQGPPPGVDL 705
>sptr|O80596|O80596 T27I1.7 protein.
Length = 1063
Score = 155 bits (391), Expect = 5e-37
Identities = 90/207 (43%), Positives = 117/207 (56%), Gaps = 13/207 (6%)
Frame = +2
Query: 20 RLXXXXEGPPPGVDLLIDSVTISYKKTASSVGGTE-------------NIILNYDFSKGL 160
R+ EGP PG DLLI SVT+ T+S TE NII N+DFS GL
Sbjct: 136 RVVLYLEGPAPGKDLLIRSVTVR-SSTSSDFQETEKNTDASNVFPLALNIIKNHDFSDGL 194
Query: 161 HPWNPIRCHAYAASQWSGFLDGIRGNSGENYAVVSKRTEHWQGLEQDITNQVSTGTVYVV 340
+ WN C ++ S L E+ AVV+ R+E WQGLEQDIT+ VS G Y V
Sbjct: 195 YSWNTNGCDSFVVSSNDCNL--------ESNAVVNNRSETWQGLEQDITDNVSPGFSYKV 246
Query: 341 SAFVRVDGIVQGQVEVKATLRLQNADGSTHYNPVGSVVASKEKWNKLEGSFSLTNMPKNV 520
SA V V G V G +V ATL+L++ +T + +G ASK+ W LEG+F ++ P V
Sbjct: 247 SASVSVSGPVLGSAQVLATLKLEHKSSATEFQLIGKTYASKDIWKTLEGTFEVSGRPDRV 306
Query: 521 VFYLEGPPAGVDLVIDSVTVACSRHKQ 601
VF+LEGPP G+DL++ SVT+ C Q
Sbjct: 307 VFFLEGPPPGIDLLVKSVTIHCESDNQ 333
Score = 137 bits (346), Expect = 9e-32
Identities = 75/166 (45%), Positives = 99/166 (59%), Gaps = 2/166 (1%)
Frame = +2
Query: 125 NIILNYDFSKGLHPWNPIRCHAYAASQ--WSGFLDGIRGNSGENYAVVSKRTEHWQGLEQ 298
NI++N DF G+ PW P C A+ S +S + +SG Y VV+ R E WQGLEQ
Sbjct: 5 NIVMNGDFFAGIEPWYPNGCEAFVVSSDPFSSEVMSADSSSG-GYVVVTNRKETWQGLEQ 63
Query: 299 DITNQVSTGTVYVVSAFVRVDGIVQGQVEVKATLRLQNADGSTHYNPVGSVVASKEKWNK 478
DIT +V++G Y VS V V G EV +T+RL++ D T Y +G AS++KW
Sbjct: 64 DITTRVASGMNYTVSTCVGVSGPFNESAEVLSTVRLEHEDSPTEYLCIGKTYASRDKWVD 123
Query: 479 LEGSFSLTNMPKNVVFYLEGPPAGVDLVIDSVTVACSRHKQSKEVK 616
LEG+FS++NMP VV YLEGP G DL+I SVTV S +E +
Sbjct: 124 LEGTFSISNMPDRVVLYLEGPAPGKDLLIRSVTVRSSTSSDFQETE 169
Score = 113 bits (283), Expect = 2e-24
Identities = 70/197 (35%), Positives = 100/197 (50%), Gaps = 10/197 (5%)
Frame = +2
Query: 20 RLXXXXEGPPPGVDLLIDSVTISY---------KKTASSVGGTENIILNYDFSKGLHPWN 172
R+ EGPPPG+DLL+ SVTI ++ S+ +I LN FS GL+ W+
Sbjct: 305 RVVFFLEGPPPGIDLLVKSVTIHCESDNQFERSREFCSAPESDNHIFLNSSFSDGLNHWS 364
Query: 173 PIRCHAYAASQWSGFLDG-IRGNSGENYAVVSKRTEHWQGLEQDITNQVSTGTVYVVSAF 349
C+ + DG I +SG +A S+RT W G+EQDIT +V +Y S+
Sbjct: 365 GRGCNLMLHESLA---DGKILPDSGTCFASASERTHKWSGIEQDITERVQRKLIYEASSV 421
Query: 350 VRVDGIVQGQVEVKATLRLQNADGSTHYNPVGSVVASKEKWNKLEGSFSLTNMPKNVVFY 529
VR+ V+ATL +Q D Y + SV + + W +L+G F L P V Y
Sbjct: 422 VRLS---HSHHTVQATLYVQYLDQREEYIGISSVQGTHDDWVELKGKFLLNGSPARAVVY 478
Query: 530 LEGPPAGVDLVIDSVTV 580
+EGPP G+D+ +D V
Sbjct: 479 IEGPPPGIDVFVDHFAV 495
Score = 68.6 bits (166), Expect = 7e-11
Identities = 55/199 (27%), Positives = 90/199 (45%), Gaps = 18/199 (9%)
Frame = +2
Query: 38 EGPPPGVDLLIDSVTI--SYKKTASSVGGTE------NIILNYDFSKG-LHPWNPIR-CH 187
EGPPPG+D+ +D + + K+T S E NI+ N S G + W P+ CH
Sbjct: 480 EGPPPGIDVFVDHFAVKPAEKETPSGRPYIESHAFGMNIVSNSHLSDGTIEGWFPLGDCH 539
Query: 188 AYAASQWSGFL-----DGIRGNSGE---NYAVVSKRTEHWQGLEQDITNQVSTGTVYVVS 343
L D +R G Y + + R+ W G Q IT++V Y VS
Sbjct: 540 LKVGDGSPRILPPLARDSLRKTQGYLSGRYVLATNRSGTWMGPAQTITDKVKLFVTYQVS 599
Query: 344 AFVRVDGIVQGQVEVKATLRLQNADGSTHYNPVGSVVASKEKWNKLEGSFSLTNMPKNVV 523
A+V++ G+ + + DG+ + G V W+++ GSF + K V+
Sbjct: 600 AWVKIGS--GGRTSPQDVNIALSVDGN--WVNGGKVEVDDGDWHEVVGSFRIEKEAKEVM 655
Query: 524 FYLEGPPAGVDLVIDSVTV 580
+++GP GVDL++ + +
Sbjct: 656 LHVQGPSPGVDLMVAGLQI 674
>sptr|Q9C643|Q9C643 Xylan endohydrolase isoenzyme, putative.
Length = 915
Score = 152 bits (383), Expect = 5e-36
Identities = 82/177 (46%), Positives = 111/177 (62%), Gaps = 2/177 (1%)
Frame = +2
Query: 101 ASSVGGTE-NIILNYDFSKGLHPWNPIRCHAYAASQWSGFLDGIRGNSG-ENYAVVSKRT 274
AS +G N+I+N+DFS G+H W+P C A+ + S G+ S +Y VV R
Sbjct: 20 ASIIGSDRTNVIVNHDFSSGMHSWHPNCCEAFVVTAESNVSHGVLDPSKCGSYVVVKNRK 79
Query: 275 EHWQGLEQDITNQVSTGTVYVVSAFVRVDGIVQGQVEVKATLRLQNADGSTHYNPVGSVV 454
E WQGLEQDITN+V ++Y VSA V V G V G VEV ATL+L++ T+Y +
Sbjct: 80 ETWQGLEQDITNRVKPCSLYKVSATVAVSGPVHGLVEVMATLKLESQQSQTNYQFIAKTC 139
Query: 455 ASKEKWNKLEGSFSLTNMPKNVVFYLEGPPAGVDLVIDSVTVACSRHKQSKEVKVPA 625
KEKW +LEG FSL ++P+ VVFYLEGP G+DL+I SVT+ H++S+ +V A
Sbjct: 140 VFKEKWVRLEGMFSLPSLPEKVVFYLEGPSPGIDLLIQSVTI----HRESEPERVTA 192
Score = 117 bits (293), Expect = 1e-25
Identities = 68/183 (37%), Positives = 103/183 (56%), Gaps = 2/183 (1%)
Frame = +2
Query: 38 EGPPPGVDLLIDSVTISYKKTASSVGGT-ENIILNYDFSKGLHPWNPIRCHAYAASQWSG 214
EGP PG+DLLI SVTI + V E I++N +F GL+ W+ C +
Sbjct: 166 EGPSPGIDLLIQSVTIHRESEPERVTAEDETIVVNPNFEDGLNNWSGRSCKIVLHDSMA- 224
Query: 215 FLDG-IRGNSGENYAVVSKRTEHWQGLEQDITNQVSTGTVYVVSAFVRVDGIVQGQVEVK 391
DG I SG+ +A ++RT++W G++Q+IT +V VY +A VR+ G V+
Sbjct: 225 --DGKIVPESGKVFASATERTQNWNGIQQEITGKVQRKRVYEATAVVRIYGNNVTTATVQ 282
Query: 392 ATLRLQNADGSTHYNPVGSVVASKEKWNKLEGSFSLTNMPKNVVFYLEGPPAGVDLVIDS 571
ATL +QN + Y + +V A+ ++W L+G F L VV Y+EGPP G D++++S
Sbjct: 283 ATLWVQNPNQRDQYIGISTVQATDKEWIHLKGKFLLNGSASRVVIYIEGPPPGTDILLNS 342
Query: 572 VTV 580
+TV
Sbjct: 343 LTV 345
Score = 70.1 bits (170), Expect = 2e-11
Identities = 51/207 (24%), Positives = 92/207 (44%), Gaps = 20/207 (9%)
Frame = +2
Query: 20 RLXXXXEGPPPGVDLLIDSVTI------------SYKKTASSVGGTENIILNYDFSKGLH 163
R+ EGPPPG D+L++S+T+ S + A V N L+ D + G
Sbjct: 324 RVVIYIEGPPPGTDILLNSLTVKHAEKIPPSPPPSIENPAFGVNILTNSHLSDDTTNG-- 381
Query: 164 PWNPI-RCHAYAASQWSGFLDGIRGNS-------GENYAVVSKRTEHWQGLEQDITNQVS 319
W + C A L + +S Y +V+ RT+ W G Q IT+++
Sbjct: 382 -WFSLGNCTLSVAEGSPRILPPMARDSLGAHERLSGRYILVTNRTQTWMGPAQMITDKLK 440
Query: 320 TGTVYVVSAFVRVDGIVQGQVEVKATLRLQNADGSTHYNPVGSVVASKEKWNKLEGSFSL 499
Y +S +V+V + V L + + + G V + ++W+++ GSF +
Sbjct: 441 LFLTYQISVWVKVGSGINSPQNVNVALGIDS-----QWVNGGQVEINDDRWHEIGGSFRI 495
Query: 500 TNMPKNVVFYLEGPPAGVDLVIDSVTV 580
P + Y++GP +G+DL++ + +
Sbjct: 496 EKNPSKALVYVQGPSSGIDLMVAGLQI 522
>sptr|Q9SM08|Q9SM08 RXF12 protein (F19C14.2 protein) (Xylan
endohydrolase).
Length = 917
Score = 149 bits (377), Expect = 2e-35
Identities = 80/171 (46%), Positives = 108/171 (63%), Gaps = 2/171 (1%)
Frame = +2
Query: 101 ASSVGGTE-NIILNYDFSKGLHPWNPIRCHAYAASQWSGFLDGIRGNSG-ENYAVVSKRT 274
AS +G N+I+N+DFS G+H W+P C A+ + S G+ S +Y VV R
Sbjct: 20 ASIIGSDRTNVIVNHDFSSGMHSWHPNCCEAFVVTAESNVSHGVLDPSKCGSYVVVKNRK 79
Query: 275 EHWQGLEQDITNQVSTGTVYVVSAFVRVDGIVQGQVEVKATLRLQNADGSTHYNPVGSVV 454
E WQGLEQDITN+V ++Y VSA V V G V G VEV ATL+L++ T+Y +
Sbjct: 80 ETWQGLEQDITNRVKPCSLYKVSATVAVSGPVHGLVEVMATLKLESQQSQTNYQFIAKTC 139
Query: 455 ASKEKWNKLEGSFSLTNMPKNVVFYLEGPPAGVDLVIDSVTVACSRHKQSK 607
KEKW +LEG FSL ++P+ VVFYLEGP G+DL+I SVT+ H++S+
Sbjct: 140 VFKEKWVRLEGMFSLPSLPEKVVFYLEGPSPGIDLLIQSVTI----HRESE 186
Score = 115 bits (289), Expect = 4e-25
Identities = 68/185 (36%), Positives = 103/185 (55%), Gaps = 4/185 (2%)
Frame = +2
Query: 38 EGPPPGVDLLIDSVTI---SYKKTASSVGGTENIILNYDFSKGLHPWNPIRCHAYAASQW 208
EGP PG+DLLI SVTI S + E I++N +F GL+ W+ C
Sbjct: 166 EGPSPGIDLLIQSVTIHRESEPELERVTAEDETIVVNPNFEDGLNNWSGRSCKIVLHDSM 225
Query: 209 SGFLDG-IRGNSGENYAVVSKRTEHWQGLEQDITNQVSTGTVYVVSAFVRVDGIVQGQVE 385
+ DG I SG+ +A ++RT++W G++Q+IT +V VY +A VR+ G
Sbjct: 226 A---DGKIVPESGKVFASATERTQNWNGIQQEITGKVQRKRVYEATAVVRIYGNNVTTAT 282
Query: 386 VKATLRLQNADGSTHYNPVGSVVASKEKWNKLEGSFSLTNMPKNVVFYLEGPPAGVDLVI 565
V+ATL +QN + Y + +V A+ ++W L+G F L VV Y+EGPP G D+++
Sbjct: 283 VQATLWVQNPNQRDQYIGISTVQATDKEWIHLKGKFLLNGSASRVVIYIEGPPPGTDILL 342
Query: 566 DSVTV 580
+S+TV
Sbjct: 343 NSLTV 347
Score = 70.1 bits (170), Expect = 2e-11
Identities = 51/207 (24%), Positives = 92/207 (44%), Gaps = 20/207 (9%)
Frame = +2
Query: 20 RLXXXXEGPPPGVDLLIDSVTI------------SYKKTASSVGGTENIILNYDFSKGLH 163
R+ EGPPPG D+L++S+T+ S + A V N L+ D + G
Sbjct: 326 RVVIYIEGPPPGTDILLNSLTVKHAEKIPPSPPPSIENPAFGVNILTNSHLSDDTTNG-- 383
Query: 164 PWNPI-RCHAYAASQWSGFLDGIRGNS-------GENYAVVSKRTEHWQGLEQDITNQVS 319
W + C A L + +S Y +V+ RT+ W G Q IT+++
Sbjct: 384 -WFSLGNCTLSVAEGSPRILPPMARDSLGAHERLSGRYILVTNRTQTWMGPAQMITDKLK 442
Query: 320 TGTVYVVSAFVRVDGIVQGQVEVKATLRLQNADGSTHYNPVGSVVASKEKWNKLEGSFSL 499
Y +S +V+V + V L + + + G V + ++W+++ GSF +
Sbjct: 443 LFLTYQISVWVKVGSGINSPQNVNVALGIDS-----QWVNGGQVEINDDRWHEIGGSFRI 497
Query: 500 TNMPKNVVFYLEGPPAGVDLVIDSVTV 580
P + Y++GP +G+DL++ + +
Sbjct: 498 EKNPSKALVYVQGPSSGIDLMVAGLQI 524
>sptrnew|AAR87197|AAR87197 Putative xylan xylanohydrolase.
Length = 756
Score = 101 bits (251), Expect = 9e-21
Identities = 63/174 (36%), Positives = 92/174 (52%), Gaps = 4/174 (2%)
Frame = +2
Query: 116 GTENIILNYDFSKGLHPWNPIRCHAYAASQWSGFLDG-IRGNSGENYAVVSKRTEHWQGL 292
G +NIILN +F GL W+ C DG + SG+ + + RT+ W G+
Sbjct: 19 GDDNIILNPEFDSGLDNWSGSGCKIELHDSLD---DGKVLAVSGKYFVAATGRTDTWNGV 75
Query: 293 EQDITNQVSTGTVYVVSAFVRVDGIVQ--GQVEVKATLRLQNADGSTHYNPVG-SVVASK 463
+QD+T+++ +Y V+A VR+ G EV+AT+ +QN DG Y V S S
Sbjct: 76 QQDVTSRLQRKLLYEVAATVRLSGAAATPSPCEVRATVAVQNTDGRQQYISVAKSPAVSD 135
Query: 464 EKWNKLEGSFSLTNMPKNVVFYLEGPPAGVDLVIDSVTVACSRHKQSKEVKVPA 625
++W +L+G F L Y+EGPPAGVDL++DS+ V +H Q K PA
Sbjct: 136 KEWVQLQGKFLLNGTVAKAAIYIEGPPAGVDLLLDSLVV---KHAQ-KATPAPA 185
Score = 81.3 bits (199), Expect = 1e-14
Identities = 65/208 (31%), Positives = 98/208 (47%), Gaps = 27/208 (12%)
Frame = +2
Query: 38 EGPPPGVDLLIDSVTISYKKTASSVGGTE--------NIILNYDFSKGLHPWNPIRCHAY 193
EGPP GVDLL+DS+ + + + A+ + NI+ N D G++ W + A
Sbjct: 159 EGPPAGVDLLLDSLVVKHAQKATPAPAPDFKNLEYGANILQNSDLDDGVNGWFGLGSCAL 218
Query: 194 A------------ASQWSGFLDGIRGNSGE----NYAVVSKRTEHWQGLEQDITNQVSTG 325
+ A Q LDG G+ E + V+ R + W G Q IT++V+
Sbjct: 219 SVHGGAPRVLPPMARQSLSPLDGDDGDGDEPLNGKHIHVTNRAQTWMGPAQVITDRVTPY 278
Query: 326 TVYVVSAFVRVDGIVQGQVEVKATLRLQNADGSTHYNPVGSVVASKEKWNKLEGSF---S 496
Y VSA+VRV G Q + + A S N G V+A E+W ++ GSF S
Sbjct: 279 ATYQVSAWVRVGG--QQAAGKPQNINVAVAVDSQWLNG-GQVMALDERWYEIGGSFRVES 335
Query: 497 LTNMPKNVVFYLEGPPAGVDLVIDSVTV 580
+ P V+ Y++GP GVDL++ + V
Sbjct: 336 SSTPPSRVMLYVQGPDPGVDLMVAGLRV 363
>sptr|Q84R74|Q84R74 Putative xylan xylanohydrolase isoenzyme.
Length = 756
Score = 101 bits (251), Expect = 9e-21
Identities = 63/174 (36%), Positives = 92/174 (52%), Gaps = 4/174 (2%)
Frame = +2
Query: 116 GTENIILNYDFSKGLHPWNPIRCHAYAASQWSGFLDG-IRGNSGENYAVVSKRTEHWQGL 292
G +NIILN +F GL W+ C DG + SG+ + + RT+ W G+
Sbjct: 19 GDDNIILNPEFDSGLDNWSGSGCKIELHDSLD---DGKVLAVSGKYFVAATGRTDTWNGV 75
Query: 293 EQDITNQVSTGTVYVVSAFVRVDGIVQ--GQVEVKATLRLQNADGSTHYNPVG-SVVASK 463
+QD+T+++ +Y V+A VR+ G EV+AT+ +QN DG Y V S S
Sbjct: 76 QQDVTSRLQRKLLYEVAATVRLSGAAATPSPCEVRATVAVQNTDGRQQYISVAKSPAVSD 135
Query: 464 EKWNKLEGSFSLTNMPKNVVFYLEGPPAGVDLVIDSVTVACSRHKQSKEVKVPA 625
++W +L+G F L Y+EGPPAGVDL++DS+ V +H Q K PA
Sbjct: 136 KEWVQLQGKFLLNGTVAKAAIYIEGPPAGVDLLLDSLVV---KHAQ-KATPAPA 185
Score = 81.3 bits (199), Expect = 1e-14
Identities = 65/208 (31%), Positives = 98/208 (47%), Gaps = 27/208 (12%)
Frame = +2
Query: 38 EGPPPGVDLLIDSVTISYKKTASSVGGTE--------NIILNYDFSKGLHPWNPIRCHAY 193
EGPP GVDLL+DS+ + + + A+ + NI+ N D G++ W + A
Sbjct: 159 EGPPAGVDLLLDSLVVKHAQKATPAPAPDFKNLEYGANILQNSDLDDGVNGWFGLGSCAL 218
Query: 194 A------------ASQWSGFLDGIRGNSGE----NYAVVSKRTEHWQGLEQDITNQVSTG 325
+ A Q LDG G+ E + V+ R + W G Q IT++V+
Sbjct: 219 SVHGGAPRVLPPMARQSLSPLDGDDGDGDEPLNGKHIHVTNRAQTWMGPAQVITDRVTPY 278
Query: 326 TVYVVSAFVRVDGIVQGQVEVKATLRLQNADGSTHYNPVGSVVASKEKWNKLEGSF---S 496
Y VSA+VRV G Q + + A S N G V+A E+W ++ GSF S
Sbjct: 279 ATYQVSAWVRVGG--QQAAGKPQNINVAVAVDSQWLNG-GQVMALDERWYEIGGSFRVES 335
Query: 497 LTNMPKNVVFYLEGPPAGVDLVIDSVTV 580
+ P V+ Y++GP GVDL++ + V
Sbjct: 336 SSTPPSRVMLYVQGPDPGVDLMVAGLRV 363
>sptr|Q9C5T7|Q9C5T7 Putative xylanase containing two cellulose
binding domains (Fragment).
Length = 232
Score = 89.0 bits (219), Expect = 5e-17
Identities = 52/154 (33%), Positives = 84/154 (54%), Gaps = 1/154 (0%)
Frame = +2
Query: 122 ENIILNYDFSKGLHPWNPIRCHAYA-ASQWSGFLDGIRGNSGENYAVVSKRTEHWQGLEQ 298
E IILN +F GL+ W C S SG + + SG+ +A ++R + W G++Q
Sbjct: 25 EKIILNPNFEDGLNNWTGRACKIVLHESMDSGKIVPL---SGKVFAAATQRKDTWNGIQQ 81
Query: 299 DITNQVSTGTVYVVSAFVRVDGIVQGQVEVKATLRLQNADGSTHYNPVGSVVASKEKWNK 478
+I+ + VY V+A VR+ G V+ATL + NA+ Y + +V A+ + W +
Sbjct: 82 EISGRFRRKRVYEVTAVVRIFGNNVTSATVQATLWVLNANKREQYIVIANVQATDKNWVE 141
Query: 479 LEGSFSLTNMPKNVVFYLEGPPAGVDLVIDSVTV 580
L+G F + P V+ YLEGPP D++++S+ V
Sbjct: 142 LKGKFVIHGSPSRVILYLEGPPPRADILLNSLVV 175
>sptr|Q8GYW5|Q8GYW5 Putative xylan endohydrolase (At4g08160).
Length = 752
Score = 89.0 bits (219), Expect = 5e-17
Identities = 52/154 (33%), Positives = 84/154 (54%), Gaps = 1/154 (0%)
Frame = +2
Query: 122 ENIILNYDFSKGLHPWNPIRCHAYA-ASQWSGFLDGIRGNSGENYAVVSKRTEHWQGLEQ 298
E IILN +F GL+ W C S SG + + SG+ +A ++R + W G++Q
Sbjct: 25 EKIILNPNFEDGLNNWTGRACKIVLHESMNSGKIVPL---SGKVFAAATQRKDTWNGIQQ 81
Query: 299 DITNQVSTGTVYVVSAFVRVDGIVQGQVEVKATLRLQNADGSTHYNPVGSVVASKEKWNK 478
+I+ + VY V+A VR+ G V+ATL + NA+ Y + +V A+ + W +
Sbjct: 82 EISGRFRRKRVYEVTAVVRIFGNNVTSATVQATLWVLNANKREQYIVIANVQATDKNWVE 141
Query: 479 LEGSFSLTNMPKNVVFYLEGPPAGVDLVIDSVTV 580
L+G F + P V+ YLEGPP D++++S+ V
Sbjct: 142 LKGKFVIHGSPSRVILYLEGPPPRADILLNSLVV 175
Score = 78.6 bits (192), Expect = 6e-14
Identities = 55/204 (26%), Positives = 95/204 (46%), Gaps = 17/204 (8%)
Frame = +2
Query: 20 RLXXXXEGPPPGVDLLIDSVTISYKKTA--------SSVGGTENIILNYD-FSKGLHPWN 172
R+ EGPPP D+L++S+ + + K + G NI+ N + G PW
Sbjct: 154 RVILYLEGPPPRADILLNSLVVQHAKRNRPSPPPFYENPGFGVNIVENSEVLDGGTKPWF 213
Query: 173 PI-RCHAYAASQWSGFLDGIRGNS-------GENYAVVSKRTEHWQGLEQDITNQVSTGT 328
+ C L + ++ G NY VV+ RT+ W G Q IT+++
Sbjct: 214 TLGNCKLSVGQGAPRTLPPMARDTLGPHKPLGGNYIVVTNRTQTWMGPAQMITDKIKLFL 273
Query: 329 VYVVSAFVRVDGIVQGQVEVKATLRLQNADGSTHYNPVGSVVASKEKWNKLEGSFSLTNM 508
Y +SA+V++ V G + + + + N V + W+++ GSF L
Sbjct: 274 TYQISAWVKLGVGVSGSSMSPQNVNIALSVDNQWVNGGQVEVTVGDTWHEIAGSFRLEKQ 333
Query: 509 PKNVVFYLEGPPAGVDLVIDSVTV 580
P+NV+ Y++GP AG+DL+I ++ +
Sbjct: 334 PQNVMVYVQGPGAGIDLMIAALQI 357
>sptrnew|CAE51307|CAE51307 Beta-1,4-xylanase.
Length = 639
Score = 71.6 bits (174), Expect = 8e-12
Identities = 39/120 (32%), Positives = 62/120 (51%), Gaps = 1/120 (0%)
Frame = +2
Query: 224 GIRGNSGENYAVVSKRTEHWQGLEQDITNQVSTGTVYVVSAFVRVDGIVQGQVEVKATLR 403
G +SG V+ RT+ W G+ QD+ ++++ G Y VSA+V+V G G +VK ++
Sbjct: 57 GTEAHSGNYSVFVTDRTQDWNGVAQDMLDKLTVGMTYQVSAWVKVAG--TGSHQVKIPMK 114
Query: 404 LQNADGSTHYNPVGSVVASKEKWNKLEGSFSLTNM-PKNVVFYLEGPPAGVDLVIDSVTV 580
Y+ + S+ +W +L G +S T N+ Y+EGP GV +D VTV
Sbjct: 115 KVETGKEPVYDNIPSITVEGSEWYRLSGPYSYTGTNVTNLELYIEGPQPGVSYYVDDVTV 174
>sptr|Q9SYE3|Q9SYE3 Putative xylan endohydrolase.
Length = 520
Score = 68.9 bits (167), Expect = 5e-11
Identities = 36/113 (31%), Positives = 62/113 (54%)
Frame = +2
Query: 242 GENYAVVSKRTEHWQGLEQDITNQVSTGTVYVVSAFVRVDGIVQGQVEVKATLRLQNADG 421
G NY VV+ RT+ W G Q IT+++ Y +SA+V++ V G + + +
Sbjct: 13 GGNYIVVTNRTQTWMGPAQMITDKIKLFLTYQISAWVKLGVGVSGSSMSPQNVNIALSVD 72
Query: 422 STHYNPVGSVVASKEKWNKLEGSFSLTNMPKNVVFYLEGPPAGVDLVIDSVTV 580
+ N V + W+++ GSF L P+NV+ Y++GP AG+DL+I ++ +
Sbjct: 73 NQWVNGGQVEVTVGDTWHEIAGSFRLEKQPQNVMVYVQGPGAGIDLMIAALQI 125
>sptr|O52373|O52373 Family 10 xylanase (EC 3.2.1.8).
Length = 1595
Score = 62.0 bits (149), Expect = 6e-09
Identities = 38/117 (32%), Positives = 61/117 (52%), Gaps = 2/117 (1%)
Frame = +2
Query: 236 NSGENYAVVSKRTEHWQGLEQDITNQVSTGTVYVVSAFVRVDGIVQGQVEVKATLRLQNA 415
+SG VS R + WQG D+TN + G Y S +V + + E+ T++ +NA
Sbjct: 229 HSGSKSLYVSGRADTWQGARIDMTNLLEKGKDYQFSIWVYQNS--GSEQEITLTMQRKNA 286
Query: 416 DGSTHYNPVG--SVVASKEKWNKLEGSFSLTNMPKNVVFYLEGPPAGVDLVIDSVTV 580
D ST Y+ + VAS W ++ GS+++ ++FY+E P A +D +D TV
Sbjct: 287 DDSTKYDTIKWRQKVASGV-WTEVSGSYTVPQTATQLIFYVESPNATLDFYLDDFTV 342
Score = 60.5 bits (145), Expect = 2e-08
Identities = 38/139 (27%), Positives = 66/139 (47%), Gaps = 1/139 (0%)
Frame = +2
Query: 218 LDGIRGNSGENYAVVSKRTEHWQGLEQDITNQVSTGTVYVVSAFVRVDGIVQGQVEVKAT 397
+D + +S VS R + WQG + D+TN + G Y S +V + ++ T
Sbjct: 380 VDNVYYHSETKALYVSGRADTWQGAQIDMTNLLEKGKDYQFSIWVYQNS--GSDQKITLT 437
Query: 398 LRLQNADGSTHYNPVG-SVVASKEKWNKLEGSFSLTNMPKNVVFYLEGPPAGVDLVIDSV 574
++ +NAD ST+Y+ + W ++ GS+++ ++FY+E P A +D +D
Sbjct: 438 MQRKNADDSTNYDTIKWQQTVPSGVWTEVSGSYTVPQTATQLIFYVESPNATLDFYLDDF 497
Query: 575 TVACSRHKQSKEVKVPARA 631
TV V +PA A
Sbjct: 498 TVI-----DKNPVTIPAAA 511
Score = 41.6 bits (96), Expect = 0.009
Identities = 25/87 (28%), Positives = 42/87 (48%)
Frame = +2
Query: 242 GENYAVVSKRTEHWQGLEQDITNQVSTGTVYVVSAFVRVDGIVQGQVEVKATLRLQNADG 421
G+ VS R+ W G D+T+ VS T+Y VS FV + + + V A ++ + +
Sbjct: 74 GDYSLKVSGRSAAWDGAIVDVTSSVSVNTMYTVSLFVYHNDVKPQRFSVYAYVKDSSGE- 132
Query: 422 STHYNPVGSVVASKEKWNKLEGSFSLT 502
Y V V + W ++ G F++T
Sbjct: 133 --RYIQVADKVVMPQYWKQIFGRFTIT 157
>sptr|Q59278|Q59278 Endoxylanase (EC 3.2.1.8).
Length = 1187
Score = 53.1 bits (126), Expect = 3e-06
Identities = 33/152 (21%), Positives = 67/152 (44%)
Frame = +2
Query: 134 LNYDFSKGLHPWNPIRCHAYAASQWSGFLDGIRGNSGENYAVVSKRTEHWQGLEQDITNQ 313
L+ DF GL W P S + G A+V+ RT+ W G+ +T+
Sbjct: 253 LSNDFEAGLGVWGPRGDATVTLSD--------DAHGGAQAALVAGRTQAWHGIGATVTDV 304
Query: 314 VSTGTVYVVSAFVRVDGIVQGQVEVKATLRLQNADGSTHYNPVGSVVASKEKWNKLEGSF 493
TG Y V A+V++ +++ +++ NA ST+ + + + W ++ +
Sbjct: 305 FQTGRTYTVDAWVKLAAGATEPADLRISVQRDNAGESTYDTVTTATGVTADAWTHVQAQY 364
Query: 494 SLTNMPKNVVFYLEGPPAGVDLVIDSVTVACS 589
++ ++ + Y+E + ++D V V+ S
Sbjct: 365 TMA-AAESALLYVESSSSLASFLVDDVVVSGS 395
>sptr|Q9AJB4|Q9AJB4 Xylanase B (EC 3.2.1.8)
(Endo-1,4-beta-xylanase).
Length = 828
Score = 52.0 bits (123), Expect = 6e-06
Identities = 41/168 (24%), Positives = 71/168 (42%), Gaps = 2/168 (1%)
Frame = +2
Query: 122 ENIILNYDFSKGLHPWNPIRCHAYAASQWSGFLDGIRGNSGENYAVVSKRTEHWQGLEQD 301
E + + F G + W+ + A+S + G VS RT++W G E +
Sbjct: 267 EGTLYSATFENGTNYWSGRGSASVASSSSKAY-------EGSKSLYVSGRTDNWNGGEVE 319
Query: 302 I-TNQVSTGTVYVVSAFVRVDGIVQGQVEVKATLRLQNADGSTHYNPVGSVVASKEKWNK 478
+ T+ G Y SA V Q+ ++ + +G+T Y + + KW K
Sbjct: 320 VNTSTFKPGNSYSFSAMVNPAESTTVQLSMQ-----YDQNGTTKYTQIAEGSCTAGKWTK 374
Query: 479 LEG-SFSLTNMPKNVVFYLEGPPAGVDLVIDSVTVACSRHKQSKEVKV 619
LE SF++ + NV Y+E P + D+ +D +A + S + V
Sbjct: 375 LENTSFTIPSGASNVKMYVEAPDSLCDIYVDRAIIADKGYVASGDTPV 422
>sptr|Q9U0G1|Q9U0G1 Xylanase B precursor.
Length = 533
Score = 49.3 bits (116), Expect = 4e-05
Identities = 28/112 (25%), Positives = 53/112 (47%)
Frame = +2
Query: 233 GNSGENYAVVSKRTEHWQGLEQDITNQVSTGTVYVVSAFVRVDGIVQGQVEVKATLRLQN 412
G +G++Y V++R ++W G + D+ + + G Y+VSA+++ + ++ +
Sbjct: 65 GKTGDSYLAVTEREKNWNGAQYDLGKKCTPGGQYLVSAWLK----TPWYCNICLSMPYTD 120
Query: 413 ADGSTHYNPVGSVVASKEKWNKLEGSFSLTNMPKNVVFYLEGPPAGVDLVID 568
G HYN + VV+ + E FS+ + +V Y E D ID
Sbjct: 121 GSGEAHYNNLKCVVSQGDWVEIKEYKFSMPSGCTDVFLYFENTSGNNDFYID 172
>sw|P36917|XYNA_THESA Endo-1,4-beta-xylanase A precursor (EC
3.2.1.8) (Xylanase A) (1,4-beta-D-xylan xylanohydrolase
A).
Length = 1157
Score = 47.4 bits (111), Expect = 2e-04
Identities = 49/197 (24%), Positives = 86/197 (43%), Gaps = 9/197 (4%)
Frame = +2
Query: 17 RRLXXXXEGPPPGVDLLIDSVTISYKKTASSVGGTENIILNYDFSKGLHPWNPIRCHAYA 196
+ L E P P ++ ID V ++ + VG N+I N F G
Sbjct: 164 KTLYMYVESPDPTLEYYIDDVVVTTQNPIQ-VG---NVIANETFENG------------N 207
Query: 197 ASQWSG----FLDGIRG--NSGENYAVVSKRTEHWQGLEQDITNQVSTGTVYVVSAFVR- 355
S W G + + G +SG+ + + RT +W G D+T ++ G Y V +V+
Sbjct: 208 TSGWIGTGSSVVKAVYGVAHSGDYSLLTTGRTANWNGPSYDLTGKIVPGQQYNVDFWVKF 267
Query: 356 VDGIVQGQVEVKATLR-LQNADGSTHYNPVGSVVASKEKWNKLEGSFSLTNMP-KNVVFY 529
V+G ++KAT++ + D N +V +K +W +++GSF+L + Y
Sbjct: 268 VNG--NDTEQIKATVKATSDKDNYIQVNDFANV--NKGEWTEIKGSFTLPVADYSGISIY 323
Query: 530 LEGPPAGVDLVIDSVTV 580
+E ++ ID +V
Sbjct: 324 VESQNPTLEFYIDDFSV 340
>sptr|O32374|O32374 Xylanase precursor.
Length = 619
Score = 47.4 bits (111), Expect = 2e-04
Identities = 41/153 (26%), Positives = 62/153 (40%), Gaps = 8/153 (5%)
Frame = +2
Query: 143 DFSKGLHPWNPIRCHAYAASQWSGFLDGIRGNSGENYAVVSKRTEHWQGLEQDITNQVST 322
DF GL+ W P + + SG VS RT W G D T+ ++
Sbjct: 39 DFETGLNGWGPRGPETVELTTEEAY-------SGRYSLKVSGRTSTWNGPMVDKTDVLTL 91
Query: 323 GTVYVVSAFVRVDGIVQGQVEVKATLRLQNADGSTH-YNPVGSVVASKEKWNKLEGSFSL 499
G Y + +V+ G E + +L+LQ DG+ Y + + K W LEG ++
Sbjct: 92 GESYKLGVYVKFVGDSYSN-EQRFSLQLQYNDGAGDVYQNIKTATVYKGTWTLLEGQLTV 150
Query: 500 TNMPKNVVFYLE-------GPPAGVDLVIDSVT 577
+ K+V Y+E P +D ID T
Sbjct: 151 PSHAKDVKIYVETEFKNSPSPQDLMDFYIDDFT 183
>sptr|Q943R0|Q943R0 Putative endo-1,4-beta-xylanase X-1.
Length = 557
Score = 47.0 bits (110), Expect = 2e-04
Identities = 29/123 (23%), Positives = 57/123 (46%), Gaps = 1/123 (0%)
Frame = +2
Query: 251 YAVVSKRTEHWQGLEQDITNQVSTGTVYVVSAFVRVDGIVQGQVEVKATLRLQNADGSTH 430
Y + + R GL + + + Y V+ ++ + +G V+ LRL + D
Sbjct: 63 YILAAGRAGEEDGLRRAVAGALKPRVTYRVAGWISLGDGAEGSHPVRVNLRLDDDDECVV 122
Query: 431 YNPVGSVVASKEKWNKLEGSFSLTNMPKNVVFYLEGPPAGVDL-VIDSVTVACSRHKQSK 607
G+V A +W +++G+F L P +++G P GVD+ V+D A R + +
Sbjct: 123 EG--GAVCAQAGRWTEIKGAFRLKASPCGATVFVQGAPDGVDVKVMDLQIFATDRRARFR 180
Query: 608 EVK 616
+++
Sbjct: 181 KLR 183
>sptr|Q60046|Q60046 XynA precursor (EC 3.2.1.8).
Length = 1234
Score = 46.6 bits (109), Expect = 3e-04
Identities = 51/197 (25%), Positives = 85/197 (43%), Gaps = 9/197 (4%)
Frame = +2
Query: 17 RRLXXXXEGPPPGVDLLIDSVTISYKKTASSVGGTENIILNYDFSKGLHPWNPIRCHAYA 196
+ L E P P ++ ID V ++ + VG N+I N F G
Sbjct: 163 KTLYMYVESPDPTLEYYIDDVVVT-PQNPIQVG---NVITNGTFENG------------N 206
Query: 197 ASQWSG----FLDGIRG--NSGENYAVVSKRTEHWQGLEQDITNQVSTGTVYVVSAFVR- 355
S W G + + G +SG + + RT +W G D+T ++ G Y V +V+
Sbjct: 207 TSGWVGTGSSVVKAVYGVAHSGGYSLLTTGRTANWNGPSYDLTGKIVPGQQYNVDFWVKF 266
Query: 356 VDGIVQGQVEVKATLR-LQNADGSTHYNPVGSVVASKEKWNKLEGSFSL-TNMPKNVVFY 529
V+G ++KAT++ N D N +V +K +W +++GSF+L V Y
Sbjct: 267 VNG--NDTEQIKATVKATSNKDNYIQVNDFVNV--NKGEWTEIKGSFTLPVTDYSGVSIY 322
Query: 530 LEGPPAGVDLVIDSVTV 580
+E ++ ID +V
Sbjct: 323 VESQNPTLEFYIDDFSV 339
>sptr|Q52644|Q52644 XynA (EC 3.2.1.8) (Endo-1,4-beta-xylanase).
Length = 680
Score = 46.2 bits (108), Expect = 4e-04
Identities = 31/117 (26%), Positives = 56/117 (47%), Gaps = 3/117 (2%)
Frame = +2
Query: 236 NSGENYAVVSKRTEHWQGLEQDITNQVSTGTVYVVSAF-VRVDGIVQGQVE--VKATLRL 406
+ G VS RT++W G ++ +GT S + + D + + E +K T++
Sbjct: 320 SEGRGSLFVSGRTDNWNGASIEL----DSGTFKQGSTYSFKTDAMQKSGKEATLKFTMQY 375
Query: 407 QNADGSTHYNPVGSVVASKEKWNKLEGSFSLTNMPKNVVFYLEGPPAGVDLVIDSVT 577
ADG HY+ + A +W LEG +++ + +V Y+E P + D ID+ +
Sbjct: 376 TAADGD-HYDQIAQQTAPDGEWITLEGDYTIPDDATKLVLYVESPDSLTDFYIDNAS 431
>sptr|O69230|O69230 Endo-1,4-beta-xylanase (EC 3.2.1.8).
Length = 1086
Score = 45.1 bits (105), Expect = 8e-04
Identities = 30/116 (25%), Positives = 56/116 (48%), Gaps = 3/116 (2%)
Frame = +2
Query: 242 GENYAVVSKRTEHWQGLEQDITNQVSTGTVYVVSAFVRVDGIVQGQV--EVKATLRLQNA 415
G+ + RTE W G +T+ V V +S +V+ +V G ++K T+ ++
Sbjct: 69 GKQSLQTTGRTEAWNGPSLSLTDVVHKNEVVEISGYVK---LVAGSAPPDLKFTVERRDR 125
Query: 416 DGSTHYNPVGSV-VASKEKWNKLEGSFSLTNMPKNVVFYLEGPPAGVDLVIDSVTV 580
+G T Y+ V + + +KW KL+G +S +++ YLE A ++D +
Sbjct: 126 NGDTQYDQVNAAEQVTDQKWVKLQGQYSY-EQGSSLLLYLESTDAKAAYLLDEFQI 180
>sptr|Q94G05|Q94G05 1,4-beta-D xylan xylanohydrolase.
Length = 556
Score = 44.3 bits (103), Expect = 0.001
Identities = 29/117 (24%), Positives = 57/117 (48%), Gaps = 1/117 (0%)
Frame = +2
Query: 251 YAVVSKRTEHWQGLEQDI-TNQVSTGTVYVVSAFVRVDGIVQGQVEVKATLRLQNADGST 427
Y +V+ R + +GL I + + Y V+ ++ + V+ L +++ +G+
Sbjct: 53 YVLVAGRADEEEGLRLPIPVDTLKPRLTYRVAGWISLGAARGTSHPVRIDLGVED-NGNE 111
Query: 428 HYNPVGSVVASKEKWNKLEGSFSLTNMPKNVVFYLEGPPAGVDLVIDSVTVACSRHK 598
G+V A + W+++ G+F L P++ Y+ G PAGVD+ + + V HK
Sbjct: 112 TLVECGAVCAKEGGWSEIMGAFRLRTEPRSAAVYVHGAPAGVDVKVMDLRVYPVDHK 168
>sptr|Q9L8L8|Q9L8L8 Beta-1,4-xylanase XynA precursor.
Length = 921
Score = 44.3 bits (103), Expect = 0.001
Identities = 32/116 (27%), Positives = 49/116 (42%), Gaps = 2/116 (1%)
Frame = +2
Query: 239 SGENYAVVSKRTEHWQGLEQDITNQVSTGTVYVVSAFVRVDGIVQGQVEVKATLRLQNA- 415
SG VS RTE W G DIT+ + G Y SA+V+ + G + ++ +Q
Sbjct: 69 SGAYGLSVSGRTETWHGPTLDITSYIQVGKTYQFSAWVK---LPSGSSNTRISMTMQRTM 125
Query: 416 DGSTHYNPVGSVVASKEKWNKLEGSFSLTNMPKNVVFYLEGPP-AGVDLVIDSVTV 580
+ +Y + A W +L+ + L N+ Y E P A ID V +
Sbjct: 126 QDTVYYEQIYFDTALSGNWIQLKAQYKLYEPAVNLQVYFEAPDHATQSFYIDDVRI 181
>sptr|Q94G06|Q94G06 1,4-beta-D xylan xylanohydrolase.
Length = 556
Score = 43.5 bits (101), Expect = 0.002
Identities = 29/117 (24%), Positives = 56/117 (47%), Gaps = 1/117 (0%)
Frame = +2
Query: 251 YAVVSKRTEHWQGLEQDI-TNQVSTGTVYVVSAFVRVDGIVQGQVEVKATLRLQNADGST 427
Y +V+ R + GL I + + Y V+ ++ + V+ L +++ +G+
Sbjct: 53 YVLVAGRADEEDGLRLPIPVDTLKPRLTYRVAGWISLGAARGTSHPVRIDLGVED-NGNE 111
Query: 428 HYNPVGSVVASKEKWNKLEGSFSLTNMPKNVVFYLEGPPAGVDLVIDSVTVACSRHK 598
G+V A + W+++ G+F L P++ Y+ G PAGVD+ + + V HK
Sbjct: 112 TLVECGAVCAKEGGWSEIMGAFRLRTEPRSAAVYVHGAPAGVDVKVMDLRVYPVDHK 168
>sptr|O52374|O52374 Family 10 xylanase (EC 3.2.1.8).
Length = 1779
Score = 43.1 bits (100), Expect = 0.003
Identities = 32/117 (27%), Positives = 57/117 (48%), Gaps = 3/117 (2%)
Frame = +2
Query: 239 SGENYAVVSKRTEHWQGLEQDITNQVSTGTVYVVSAFVRVDGIVQGQVEVKATLRLQNAD 418
+G++ VS R+ W G+ D+ N ++ GT +VVSA+V+ Q V + D
Sbjct: 63 NGKSSIRVSNRSSIWDGVAVDVKNIMNNGTTWVVSAYVK----HSYQKPVAFGISAVYDD 118
Query: 419 GS-THYNPVGSVVASKEKWNKLEGSF--SLTNMPKNVVFYLEGPPAGVDLVIDSVTV 580
GS +G VVA W K+ G + +++N+ ++ +GVD +D + +
Sbjct: 119 GSGVKSTLIGEVVAIPNYWKKIVGKWTPNISNVRNLLIVVHTIVESGVDYNVDYIQI 175
>sptr|Q943R2|Q943R2 Putative endo-1,4-beta-xylanase X-1.
Length = 567
Score = 42.4 bits (98), Expect = 0.005
Identities = 41/147 (27%), Positives = 63/147 (42%), Gaps = 11/147 (7%)
Frame = +2
Query: 221 DGIRGNSGENYAVVSKRTEHWQGLEQDITNQVSTGTVYVVSAFVRVDGIVQGQVEVKAT- 397
D R Y + + R GL ++++ + Y V+ +V + G A
Sbjct: 61 DEPRPRPSGRYVLAAHRAGERDGLCRELSRAPAAKVTYRVAGWVGLQGAGAADGCCHAVR 120
Query: 398 LRLQNADGSTHYNPVGS--VVASKEKWNKLEGSFSLTN--MPKNVVFYLEGPPAGVDLVI 565
+ + DG PVG VVA KW ++ GSF + + P+ ++ GPPAGVDL +
Sbjct: 121 VEVCTDDG----RPVGGGVVVAEAGKWGEIMGSFRVDDDEPPRCAKVFVHGPPAGVDLKV 176
Query: 566 DSVTV------ACSRHKQSKEVKVPAR 628
+ V A RH + K KV R
Sbjct: 177 MDLQVFAVNKIARLRHLRKKTDKVRKR 203
>sptr|Q9F1V3|Q9F1V3 Xylanase A.
Length = 1050
Score = 41.6 bits (96), Expect = 0.009
Identities = 26/113 (23%), Positives = 55/113 (48%), Gaps = 3/113 (2%)
Frame = +2
Query: 248 NYAV-VSKRTEHWQGLEQDITNQVSTGTVYVVSAFVRVDGIVQGQVEVKATLRLQNADGS 424
+YA+ V RT W G + VS G+ Y V+A+V++ I +++ + ++ N +
Sbjct: 233 SYALKVENRTMTWHGPSLHVEKNVSQGSEYKVTAWVKL--ISPESTQLQLSTQVGNGGSA 290
Query: 425 THYNPVGSVVASKEKWNKLEGSFSLTNMPKN-VVFYLE-GPPAGVDLVIDSVT 577
++ + +++ + W K EG++ N+ + Y+E A ID ++
Sbjct: 291 SYVSLAAKTISTADGWVKFEGTYRYNNVSSGYITIYVESASSANASFYIDDIS 343
>sptr|Q9I060|Q9I060 Hypothetical protein PA2783.
Length = 599
Score = 41.2 bits (95), Expect = 0.011
Identities = 36/153 (23%), Positives = 52/153 (33%)
Frame = +2
Query: 53 GVDLLIDSVTISYKKTASSVGGTENIILNYDFSKGLHPWNPIRCHAYAASQWSGFLDGIR 232
G LL+D + + + A + YDF G+ W+ + A A S
Sbjct: 429 GASLLVDEMNLQEAQAAPPSVPPAPKRIAYDFESGIGGWSGVHASARATRVAS------- 481
Query: 233 GNSGENYAVVSKRTEHWQGLEQDITNQVSTGTVYVVSAFVRVDGIVQGQVEVKATLRLQN 412
+G +R G + + G Y SA VRV Q A L L++
Sbjct: 482 --AGRLALEAYQRRYAGTGASTSLLGNLEAGRTYAFSADVRVGDGRGSQAMTYAYLYLES 539
Query: 413 ADGSTHYNPVGSVVASKEKWNKLEGSFSLTNMP 511
Y P+G V +W L G L P
Sbjct: 540 QGRPGEYLPLGYKVVENGRWASLRGQVQLPKGP 572
>sptr|Q9RQB7|Q9RQB7 Endo-1,4-beta-xylanase (EC 3.2.1.8).
Length = 1183
Score = 41.2 bits (95), Expect = 0.011
Identities = 43/151 (28%), Positives = 73/151 (48%), Gaps = 5/151 (3%)
Frame = +2
Query: 62 LLIDSVTISYKKTASSVG-GTENIILNYDFS-KGLHPWNPIRCHAYAASQWSGFLDGIRG 235
LL+D + ++ +T++ G +I + DF + L W P + A + + G G
Sbjct: 194 LLVDDIVVTTTETSTGEGPAAGTVIADTDFDDQTLQGWVPRQPDDTAPTL--AVVAG--G 249
Query: 236 NSGENYAV-VSKRTEHWQGLEQDITNQVSTGTVYVVSAFVR-VDGIVQGQVEVKATLRLQ 409
G YA VS R GL+ D+ +G A+VR DG G++ + A R
Sbjct: 250 ADGTGYAAQVSDRDSDGDGLQYDVAAAGVSGATLQYEAWVRFADGEPAGEMTLSA--RTV 307
Query: 410 NADGSTHYNPVGSVV-ASKEKWNKLEGSFSL 499
NA G+T Y+ + S+ A+ + W K+ G+F++
Sbjct: 308 NA-GATAYSNLSSISGATNDGWTKVGGTFTM 337
>sptr|Q94G07|Q94G07 1,4-beta-D xylan xylanohydrolase (Fragment).
Length = 497
Score = 40.4 bits (93), Expect = 0.019
Identities = 18/52 (34%), Positives = 30/52 (57%)
Frame = +2
Query: 443 GSVVASKEKWNKLEGSFSLTNMPKNVVFYLEGPPAGVDLVIDSVTVACSRHK 598
G+V A + W+++ G+F L P++ Y+ G PAGVD+ + + V HK
Sbjct: 58 GAVCAKEGGWSEIMGAFRLRTEPRSAAVYVHGAPAGVDVKVMDLRVYPVDHK 109
>sptr|Q9X3P5|Q9X3P5 XynA.
Length = 1770
Score = 40.4 bits (93), Expect = 0.019
Identities = 27/87 (31%), Positives = 43/87 (49%), Gaps = 1/87 (1%)
Frame = +2
Query: 239 SGENYAVVSKRTEHWQGLEQDITNQVSTGTVYVVSAFVRVDGIVQGQVEVKATLRLQNAD 418
+G++ VS R+ W G+ D+ N ++ GT +VVSA+V+ Q V + D
Sbjct: 63 NGKSSVRVSNRSSIWDGVAVDVKNIMNNGTTWVVSAYVK----HSYQKPVAFGISAVYDD 118
Query: 419 GS-THYNPVGSVVASKEKWNKLEGSFS 496
GS +G VVA W K+ G ++
Sbjct: 119 GSGVKSTLIGEVVAIPNYWKKIVGKWT 145
Database: /db/uniprot/tmp/swall
Posted date: Mar 5, 2004 7:45 PM
Number of letters in database: 442,889,342
Number of sequences in database: 1,395,590
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 514,381,173
Number of Sequences: 1395590
Number of extensions: 10737381
Number of successful extensions: 28385
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 27386
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 28331
length of database: 442,889,342
effective HSP length: 118
effective length of database: 278,209,722
effective search space used: 25873504146
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

