BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
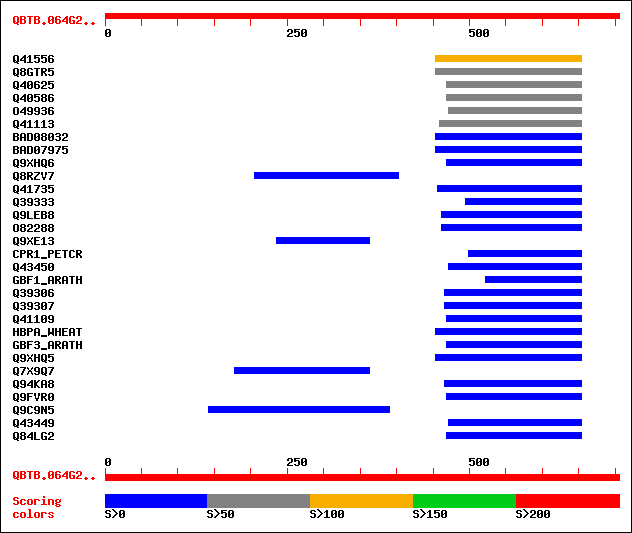
Query= QBTB.064G21F020918.3.1
(705 letters)
Database: swall
1,381,838 sequences; 439,479,560 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sptr|Q41556|Q41556 Transcription factor HBP-1a(c14). 122 4e-27
sptr|Q8GTR5|Q8GTR5 bZIP transcription factor ZIP1. 59 8e-08
sptr|Q40625|Q40625 DNA-binding factor of bZIP class. 55 7e-07
sptr|Q40586|Q40586 TAF-2. 53 5e-06
sptr|O49936|O49936 Basic leucine zipper protein. 50 2e-05
sptr|Q41113|Q41113 bZIP transcriptional repressor ROM1. 50 2e-05
sptrnew|BAD08032|BAD08032 Putative transcription factor HBP-1a. 49 5e-05
sptrnew|BAD07975|BAD07975 Putative transcription factor HBP-1a. 49 5e-05
sptr|Q9XHQ6|Q9XHQ6 G-box binding protein 1. 49 7e-05
sptr|Q8RZV7|Q8RZV7 Putative receptor-like protein (Putative leuc... 49 9e-05
sptr|Q41735|Q41735 G-box binding factor 1. 48 1e-04
sptr|Q39333|Q39333 G-box binding factor 1A. 48 1e-04
sptr|Q9LEB8|Q9LEB8 Common plant regulatory factor 5. 47 3e-04
sptr|O82288|O82288 Putative G-box-binding bZIP transcription fac... 46 4e-04
sptr|Q9XE13|Q9XE13 EST C96716(C10608) corresponds to a region of... 45 0.001
sw|Q99089|CPR1_PETCR Common plant regulatory factor CPRF-1. 44 0.003
sptr|Q43450|Q43450 G-box binding factor (Fragment). 44 0.003
sw|P42774|GBF1_ARATH G-box binding factor 1. 44 0.003
sptr|Q39306|Q39306 Transcription factor. 43 0.004
sptr|Q39307|Q39307 Transcription factor. 43 0.005
sptr|Q41109|Q41109 Regulator of MAT2. 42 0.008
sw|P23922|HBPA_WHEAT Transcription factor HBP-1a (Histone-specif... 42 0.008
sw|P42776|GBF3_ARATH G-box binding factor 3. 42 0.011
sptr|Q9XHQ5|Q9XHQ5 G-box binding protein 2. 41 0.014
sptr|Q7X9Q7|Q7X9Q7 Polygalacturonase-inhibiting protein. 41 0.018
sptr|Q94KA8|Q94KA8 bZIP transcription factor 2. 41 0.018
sptr|Q9FVR0|Q9FVR0 G-box binding protein, putative. 40 0.031
sptr|Q9C9N5|Q9C9N5 Receptor protein kinase, putative. 40 0.031
sptr|Q43449|Q43449 G-box binding factor. 40 0.031
sptr|Q84LG2|Q84LG2 Putative G-box binding protein. 40 0.031
>sptr|Q41556|Q41556 Transcription factor HBP-1a(c14).
Length = 381
Score = 122 bits (307), Expect = 4e-27
Identities = 54/67 (80%), Positives = 58/67 (86%)
Frame = +3
Query: 453 DVATRSKSQKSSAIQNEPSTPANPPTAYPDWSQFQAYYNAAGTAPVTPPAFFHSSVAPTH 632
DVATRSKSQKSS QNE STP NPPTAYPDWSQFQAYYN GT +TPPA+FHS+VAP+
Sbjct: 5 DVATRSKSQKSSTAQNEQSTPTNPPTAYPDWSQFQAYYNVPGTTQMTPPAYFHSTVAPSP 64
Query: 633 QGHPYMW 653
QGHPYMW
Sbjct: 65 QGHPYMW 71
>sptr|Q8GTR5|Q8GTR5 bZIP transcription factor ZIP1.
Length = 367
Score = 58.5 bits (140), Expect = 8e-08
Identities = 28/69 (40%), Positives = 37/69 (53%), Gaps = 2/69 (2%)
Frame = +3
Query: 453 DVATRSKSQKSSAIQNE--PSTPANPPTAYPDWSQFQAYYNAAGTAPVTPPAFFHSSVAP 626
+ T +K+ K+S Q + P++ PT YPDW+ FQ Y P+ P FF S V
Sbjct: 5 EAETPAKANKASTPQEQQPPTSSTTTPTVYPDWTNFQGY------PPIPPHGFFPSPVVS 58
Query: 627 THQGHPYMW 653
QGHPYMW
Sbjct: 59 NPQGHPYMW 67
>sptr|Q40625|Q40625 DNA-binding factor of bZIP class.
Length = 390
Score = 55.5 bits (132), Expect = 7e-07
Identities = 28/65 (43%), Positives = 36/65 (55%), Gaps = 3/65 (4%)
Frame = +3
Query: 468 SKSQKSSAIQNEP---STPANPPTAYPDWSQFQAYYNAAGTAPVTPPAFFHSSVAPTHQG 638
+K+ K+SA Q + S+ P YPDW+ FQ Y P+ P FF S VA + QG
Sbjct: 10 TKTSKASAPQEQQPPASSSTATPAVYPDWANFQGY------PPIPPHGFFPSPVASSPQG 63
Query: 639 HPYMW 653
HPYMW
Sbjct: 64 HPYMW 68
>sptr|Q40586|Q40586 TAF-2.
Length = 415
Score = 52.8 bits (125), Expect = 5e-06
Identities = 27/62 (43%), Positives = 35/62 (56%)
Frame = +3
Query: 468 SKSQKSSAIQNEPSTPANPPTAYPDWSQFQAYYNAAGTAPVTPPAFFHSSVAPTHQGHPY 647
SK +KSS+ P+ + YPDW+ QAYY V P +F+S+VAP H HPY
Sbjct: 10 SKPEKSSS----PTPDQSNLHVYPDWAAMQAYYGPR----VAVPTYFNSAVAPGHTPHPY 61
Query: 648 MW 653
MW
Sbjct: 62 MW 63
>sptr|O49936|O49936 Basic leucine zipper protein.
Length = 422
Score = 50.4 bits (119), Expect = 2e-05
Identities = 27/61 (44%), Positives = 34/61 (55%)
Frame = +3
Query: 471 KSQKSSAIQNEPSTPANPPTAYPDWSQFQAYYNAAGTAPVTPPAFFHSSVAPTHQGHPYM 650
KS+K+S P+T N YPDW+ QAYY V P +F+S+VAP H PYM
Sbjct: 11 KSEKTSP----PATEHNGVHMYPDWAAMQAYYGPR----VALPPYFNSAVAPGHPPPPYM 62
Query: 651 W 653
W
Sbjct: 63 W 63
>sptr|Q41113|Q41113 bZIP transcriptional repressor ROM1.
Length = 339
Score = 50.4 bits (119), Expect = 2e-05
Identities = 30/66 (45%), Positives = 37/66 (56%), Gaps = 1/66 (1%)
Frame = +3
Query: 459 ATRSKSQKSSAIQNEPSTPANPPTAYPDWSQFQAYYNAAGTAPVTPPAFFHSSVA-PTHQ 635
+T S+ SS++Q P+ PA YPDWS Y A G A PP FF S+VA PT
Sbjct: 7 STTKSSKSSSSVQETPTVPA-----YPDWSSSMQAYYAPGAA---PPPFFASTVASPT-- 56
Query: 636 GHPYMW 653
HPY+W
Sbjct: 57 PHPYLW 62
>sptrnew|BAD08032|BAD08032 Putative transcription factor HBP-1a.
Length = 347
Score = 49.3 bits (116), Expect = 5e-05
Identities = 26/68 (38%), Positives = 35/68 (51%), Gaps = 1/68 (1%)
Frame = +3
Query: 453 DVATRSKSQKSSAIQNEPSTPANPPT-AYPDWSQFQAYYNAAGTAPVTPPAFFHSSVAPT 629
D T SK+ K+S + P+T + YP+W FQAY + + P FF VA +
Sbjct: 5 DPGTPSKATKASEPEQSPATTSGTTAPVYPEWPGFQAY------SAIPPHGFFPPPVAAS 58
Query: 630 HQGHPYMW 653
Q HPYMW
Sbjct: 59 PQAHPYMW 66
>sptrnew|BAD07975|BAD07975 Putative transcription factor HBP-1a.
Length = 347
Score = 49.3 bits (116), Expect = 5e-05
Identities = 26/68 (38%), Positives = 35/68 (51%), Gaps = 1/68 (1%)
Frame = +3
Query: 453 DVATRSKSQKSSAIQNEPSTPANPPT-AYPDWSQFQAYYNAAGTAPVTPPAFFHSSVAPT 629
D T SK+ K+S + P+T + YP+W FQAY + + P FF VA +
Sbjct: 5 DPGTPSKATKASEPEQSPATTSGTTAPVYPEWPGFQAY------SAIPPHGFFPPPVAAS 58
Query: 630 HQGHPYMW 653
Q HPYMW
Sbjct: 59 PQAHPYMW 66
>sptr|Q9XHQ6|Q9XHQ6 G-box binding protein 1.
Length = 426
Score = 48.9 bits (115), Expect = 7e-05
Identities = 26/62 (41%), Positives = 33/62 (53%)
Frame = +3
Query: 468 SKSQKSSAIQNEPSTPANPPTAYPDWSQFQAYYNAAGTAPVTPPAFFHSSVAPTHQGHPY 647
SK +KSS+ P+ + YPDW+ QAYY V P +F S+VA H HPY
Sbjct: 10 SKPEKSSS----PAPEQSNVHVYPDWAAMQAYYGPR----VAVPPYFSSAVASGHPPHPY 61
Query: 648 MW 653
MW
Sbjct: 62 MW 63
>sptr|Q8RZV7|Q8RZV7 Putative receptor-like protein (Putative
leucin-rich repeat protein kinase).
Length = 1294
Score = 48.5 bits (114), Expect = 9e-05
Identities = 23/41 (56%), Positives = 28/41 (68%)
Frame = +1
Query: 238 LNLSFNAFTGSIPEEVAELEAIAVFFN*REQIFGSYPDWIR 360
+NLSFNA G IPEE A+LEAI FF ++ G PDWI+
Sbjct: 358 INLSFNALIGPIPEEFADLEAIVSFFVEGNKLSGRVPDWIQ 398
Score = 38.5 bits (88), Expect = 0.090
Identities = 21/66 (31%), Positives = 36/66 (54%)
Frame = +1
Query: 205 PGLVSSSAG*TLNLSFNAFTGSIPEEVAELEAIAVFFN*REQIFGSYPDWIRNM*KLSFW 384
PG+ S + TL+LS N+F G+IP E+ +LE + + + + G P I ++ +L
Sbjct: 227 PGITSLTNLLTLDLSSNSFEGTIPREIGQLENLELLILGKNDLTGRIPQEIGSLKQLKLL 286
Query: 385 DLKRIQ 402
L+ Q
Sbjct: 287 HLEECQ 292
>sptr|Q41735|Q41735 G-box binding factor 1.
Length = 377
Score = 48.1 bits (113), Expect = 1e-04
Identities = 26/66 (39%), Positives = 35/66 (53%)
Frame = +3
Query: 456 VATRSKSQKSSAIQNEPSTPANPPTAYPDWSQFQAYYNAAGTAPVTPPAFFHSSVAPTHQ 635
VAT+ +S ++ P+ P+ YPDWS QAYY V PP +F ++AP H
Sbjct: 7 VATQKTGNTTSPSKDYPT-----PSPYPDWSTMQAYYGPG----VLPPTYFTPAIAPGHP 57
Query: 636 GHPYMW 653
PYMW
Sbjct: 58 -PPYMW 62
>sptr|Q39333|Q39333 G-box binding factor 1A.
Length = 313
Score = 47.8 bits (112), Expect = 1e-04
Identities = 28/59 (47%), Positives = 31/59 (52%), Gaps = 6/59 (10%)
Frame = +3
Query: 495 QNEPSTPAN-----PPTAYPDWS-QFQAYYNAAGTAPVTPPAFFHSSVAPTHQGHPYMW 653
+ PS PA+ PPT YPDWS QAYY G TP FF S V + HPYMW
Sbjct: 6 EKTPSKPASSTQDIPPTPYPDWSNSMQAYYGGGG----TPSPFFPSPVG-SPSPHPYMW 59
>sptr|Q9LEB8|Q9LEB8 Common plant regulatory factor 5.
Length = 352
Score = 46.6 bits (109), Expect = 3e-04
Identities = 30/65 (46%), Positives = 34/65 (52%), Gaps = 1/65 (1%)
Frame = +3
Query: 462 TRSKSQKSSAIQNEPSTPANPPTAYPDW-SQFQAYYNAAGTAPVTPPAFFHSSVAPTHQG 638
T SK K +A E TP + YPDW S QAYY A PAF+ S+VAP
Sbjct: 8 TPSKHSKPTAPVQEVQTPPS----YPDWSSSMQAYYGAGAA-----PAFYASTVAPP-TP 57
Query: 639 HPYMW 653
HPYMW
Sbjct: 58 HPYMW 62
>sptr|O82288|O82288 Putative G-box-binding bZIP transcription
factor.
Length = 368
Score = 46.2 bits (108), Expect = 4e-04
Identities = 26/64 (40%), Positives = 32/64 (50%)
Frame = +3
Query: 462 TRSKSQKSSAIQNEPSTPANPPTAYPDWSQFQAYYNAAGTAPVTPPAFFHSSVAPTHQGH 641
T S + EPS+ + A PDWS FQAY +P+ PP H VA + Q H
Sbjct: 17 TPPPSSTAPPSSQEPSSAVSAGMATPDWSGFQAY------SPMPPP---HGYVASSPQPH 67
Query: 642 PYMW 653
PYMW
Sbjct: 68 PYMW 71
>sptr|Q9XE13|Q9XE13 EST C96716(C10608) corresponds to a region of
the predicted gene.
Length = 1420
Score = 45.1 bits (105), Expect = 0.001
Identities = 22/43 (51%), Positives = 30/43 (69%)
Frame = +1
Query: 235 TLNLSFNAFTGSIPEEVAELEAIAVFFN*REQIFGSYPDWIRN 363
TL LS N FTG+IPEE+A+L A+ +F ++ G PDWI+N
Sbjct: 461 TLVLSGNNFTGTIPEELADLVAVVLFDVEGNRLSGHIPDWIQN 503
>sw|Q99089|CPR1_PETCR Common plant regulatory factor CPRF-1.
Length = 411
Score = 43.5 bits (101), Expect = 0.003
Identities = 22/56 (39%), Positives = 28/56 (50%), Gaps = 4/56 (7%)
Frame = +3
Query: 498 NEPSTPANPPTA----YPDWSQFQAYYNAAGTAPVTPPAFFHSSVAPTHQGHPYMW 653
+ P PA P + YPDW+ QAYY V P +F+ +VA HPYMW
Sbjct: 16 SSPPPPAAPDQSNSHVYPDWAAMQAYYGPR----VALPPYFNPAVASGQSPHPYMW 67
>sptr|Q43450|Q43450 G-box binding factor (Fragment).
Length = 423
Score = 43.5 bits (101), Expect = 0.003
Identities = 26/65 (40%), Positives = 34/65 (52%), Gaps = 4/65 (6%)
Frame = +3
Query: 471 KSQKSSAIQNEPSTP--ANPPT--AYPDWSQFQAYYNAAGTAPVTPPAFFHSSVAPTHQG 638
KS K+ + + P+T N P YPDW+ Q YY V P +F+S+VA H
Sbjct: 7 KSVKTGSPSSSPATTEQTNQPNIHVYPDWAAMQ-YYGPR----VNIPPYFNSAVASGHAP 61
Query: 639 HPYMW 653
HPYMW
Sbjct: 62 HPYMW 66
>sw|P42774|GBF1_ARATH G-box binding factor 1.
Length = 315
Score = 43.5 bits (101), Expect = 0.003
Identities = 23/45 (51%), Positives = 25/45 (55%), Gaps = 1/45 (2%)
Frame = +3
Query: 522 PPTAYPDW-SQFQAYYNAAGTAPVTPPAFFHSSVAPTHQGHPYMW 653
PPT YPDW + QAYY G TP FF S V + HPYMW
Sbjct: 23 PPTPYPDWQNSMQAYYGGGG----TPNPFFPSPVG-SPSPHPYMW 62
>sptr|Q39306|Q39306 Transcription factor.
Length = 376
Score = 43.1 bits (100), Expect = 0.004
Identities = 24/68 (35%), Positives = 35/68 (51%), Gaps = 5/68 (7%)
Frame = +3
Query: 465 RSKSQKSSAIQNEPSTPANPPT---AYPDWSQFQAYYNAAGTAPVTPPAFFHSSVAPT-- 629
+S+ K + N+PS+P T YPDW+ QAYY V P +++S++A
Sbjct: 3 KSEEPKVTKSDNKPSSPPADQTNVHVYPDWAAMQAYYGPR----VAMPPYYNSAMAAASG 58
Query: 630 HQGHPYMW 653
H PYMW
Sbjct: 59 HPPPPYMW 66
>sptr|Q39307|Q39307 Transcription factor.
Length = 374
Score = 42.7 bits (99), Expect = 0.005
Identities = 24/68 (35%), Positives = 35/68 (51%), Gaps = 5/68 (7%)
Frame = +3
Query: 465 RSKSQKSSAIQNEPSTPANPPT---AYPDWSQFQAYYNAAGTAPVTPPAFFHSSVAPT-- 629
+S+ K + N+PS+P T YPDW+ QAYY V P +++S++A
Sbjct: 3 KSEEPKVTKSDNKPSSPPADQTNVHVYPDWAAMQAYYGPR----VAIPPYYNSAMAAASG 58
Query: 630 HQGHPYMW 653
H PYMW
Sbjct: 59 HPPPPYMW 66
>sptr|Q41109|Q41109 Regulator of MAT2.
Length = 424
Score = 42.0 bits (97), Expect = 0.008
Identities = 26/70 (37%), Positives = 33/70 (47%), Gaps = 8/70 (11%)
Frame = +3
Query: 468 SKSQKSSAIQNEPSTPA------NPPT--AYPDWSQFQAYYNAAGTAPVTPPAFFHSSVA 623
+ + S PS+PA N P YPDW+ Q YY V P +F+S+VA
Sbjct: 3 NSEEGKSVKTGSPSSPATTTNQTNQPNFHVYPDWAAMQ-YYGPR----VNIPPYFNSAVA 57
Query: 624 PTHQGHPYMW 653
H HPYMW
Sbjct: 58 SGHAPHPYMW 67
>sw|P23922|HBPA_WHEAT Transcription factor HBP-1a (Histone-specific
transcription factor HBP1).
Length = 349
Score = 42.0 bits (97), Expect = 0.008
Identities = 23/68 (33%), Positives = 29/68 (42%), Gaps = 1/68 (1%)
Frame = +3
Query: 453 DVATRSKSQKSSAIQNEPSTPANPPT-AYPDWSQFQAYYNAAGTAPVTPPAFFHSSVAPT 629
D +T SK+ K + P+T + YP+W FQ Y P PP F
Sbjct: 5 DPSTPSKASKPPEQEQPPATTSGTTAPVYPEWPGFQGY-------PAMPPHGFFPPPVAA 57
Query: 630 HQGHPYMW 653
Q HPYMW
Sbjct: 58 GQAHPYMW 65
>sw|P42776|GBF3_ARATH G-box binding factor 3.
Length = 382
Score = 41.6 bits (96), Expect = 0.011
Identities = 23/65 (35%), Positives = 30/65 (46%), Gaps = 3/65 (4%)
Frame = +3
Query: 468 SKSQKSSAIQNEPSTPANPPT---AYPDWSQFQAYYNAAGTAPVTPPAFFHSSVAPTHQG 638
S+ K ++PS+P T YPDW+ QAYY G PP + + A H
Sbjct: 5 SEEPKPPTKSDKPSSPPVDQTNVHVYPDWAAMQAYY---GPRVAMPPYYNSAMAASGHPP 61
Query: 639 HPYMW 653
PYMW
Sbjct: 62 PPYMW 66
>sptr|Q9XHQ5|Q9XHQ5 G-box binding protein 2.
Length = 394
Score = 41.2 bits (95), Expect = 0.014
Identities = 23/70 (32%), Positives = 34/70 (48%), Gaps = 3/70 (4%)
Frame = +3
Query: 453 DVATRSKSQKSSAIQNEPST---PANPPTAYPDWSQFQAYYNAAGTAPVTPPAFFHSSVA 623
D +++ + A + PS+ PA PDW+ FQAY +P+ P H +A
Sbjct: 7 DKSSKEAKEAKEAKETPPSSQEQPAATSAGTPDWTGFQAY------SPIPP----HGFLA 56
Query: 624 PTHQGHPYMW 653
+ Q HPYMW
Sbjct: 57 SSPQAHPYMW 66
>sptr|Q7X9Q7|Q7X9Q7 Polygalacturonase-inhibiting protein.
Length = 326
Score = 40.8 bits (94), Expect = 0.018
Identities = 26/64 (40%), Positives = 33/64 (51%), Gaps = 2/64 (3%)
Frame = +1
Query: 178 LAWSG--DPPPPGLVSSSAG*TLNLSFNAFTGSIPEEVAELEAIAVFFN*REQIFGSYPD 351
L+W+G P P L S S L+LSFN FTGSIP +A L + R ++ G PD
Sbjct: 122 LSWNGLSGPIPSFLGSLSNLDILDLSFNRFTGSIPSSLANLRRLGTLHLDRNKLTGPIPD 181
Query: 352 WIRN 363
N
Sbjct: 182 SFGN 185
>sptr|Q94KA8|Q94KA8 bZIP transcription factor 2.
Length = 417
Score = 40.8 bits (94), Expect = 0.018
Identities = 24/68 (35%), Positives = 32/68 (47%), Gaps = 5/68 (7%)
Frame = +3
Query: 465 RSKSQKSSAIQNEPSTPANPPTA-----YPDWSQFQAYYNAAGTAPVTPPAFFHSSVAPT 629
++ +K S S +PPT PDWS FQAY +P+ P H +A +
Sbjct: 8 KTPKEKESKTPPATSQEQSPPTTGMATINPDWSNFQAY------SPMPP----HGFLASS 57
Query: 630 HQGHPYMW 653
Q HPYMW
Sbjct: 58 PQAHPYMW 65
>sptr|Q9FVR0|Q9FVR0 G-box binding protein, putative.
Length = 387
Score = 40.0 bits (92), Expect = 0.031
Identities = 24/63 (38%), Positives = 32/63 (50%), Gaps = 1/63 (1%)
Frame = +3
Query: 468 SKSQKSSAIQNEPSTPANPPTAYP-DWSQFQAYYNAAGTAPVTPPAFFHSSVAPTHQGHP 644
S S ++ + EPS+ + A DWS FQAY +P+ P H VA + Q HP
Sbjct: 23 SSSAPATVVSQEPSSAVSAGVAVTQDWSGFQAY------SPMPP----HGYVASSPQPHP 72
Query: 645 YMW 653
YMW
Sbjct: 73 YMW 75
>sptr|Q9C9N5|Q9C9N5 Receptor protein kinase, putative.
Length = 685
Score = 40.0 bits (92), Expect = 0.031
Identities = 32/85 (37%), Positives = 46/85 (54%), Gaps = 2/85 (2%)
Frame = +1
Query: 142 PSPRLVFLLSAVLAWSGDPPPPGLVSSSAG*-TLNLSFNAFTGSIPEEVAELEAIAVFFN 318
P +L L+ + ++SGD P GL S+ TLNLSFN TG+IPE+V LE + +
Sbjct: 160 PCKKLKTLVLSKNSFSGDLPT-GLGSNLVHLRTLNLSFNRLTGTIPEDVGSLENLKGTLD 218
Query: 319 *REQIF-GSYPDWIRNM*KLSFWDL 390
F G P + N+ +L + DL
Sbjct: 219 LSHNFFSGMIPTSLGNLPELLYVDL 243
>sptr|Q43449|Q43449 G-box binding factor.
Length = 341
Score = 40.0 bits (92), Expect = 0.031
Identities = 28/62 (45%), Positives = 33/62 (53%), Gaps = 1/62 (1%)
Frame = +3
Query: 471 KSQKSSAIQNEPSTPANPPTAYPDWSQFQAYYNAAGTAPVTPPAFFHSSVA-PTHQGHPY 647
K +S+IQ P P +YPDWS Y A G TPPAFF S++A PT H Y
Sbjct: 13 KPSSTSSIQ----IPLAP--SYPDWSSSMQAYYAPG---ATPPAFFASNIASPT--PHSY 61
Query: 648 MW 653
MW
Sbjct: 62 MW 63
>sptr|Q84LG2|Q84LG2 Putative G-box binding protein.
Length = 389
Score = 40.0 bits (92), Expect = 0.031
Identities = 24/63 (38%), Positives = 32/63 (50%), Gaps = 1/63 (1%)
Frame = +3
Query: 468 SKSQKSSAIQNEPSTPANPPTAYP-DWSQFQAYYNAAGTAPVTPPAFFHSSVAPTHQGHP 644
S S ++ + EPS+ + A DWS FQAY +P+ P H VA + Q HP
Sbjct: 23 SSSAPATVVSQEPSSAVSAGVAVTQDWSGFQAY------SPMPP----HGYVASSPQPHP 72
Query: 645 YMW 653
YMW
Sbjct: 73 YMW 75
Database: swall
Posted date: Feb 26, 2004 3:00 PM
Number of letters in database: 439,479,560
Number of sequences in database: 1,381,838
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 602,634,800
Number of Sequences: 1381838
Number of extensions: 13473416
Number of successful extensions: 48859
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 43448
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 48626
length of database: 439,479,560
effective HSP length: 119
effective length of database: 275,040,838
effective search space used: 31629696370
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

