BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= QBTB.064D08F020918.3.1
(735 letters)
Database: swall
1,381,838 sequences; 439,479,560 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
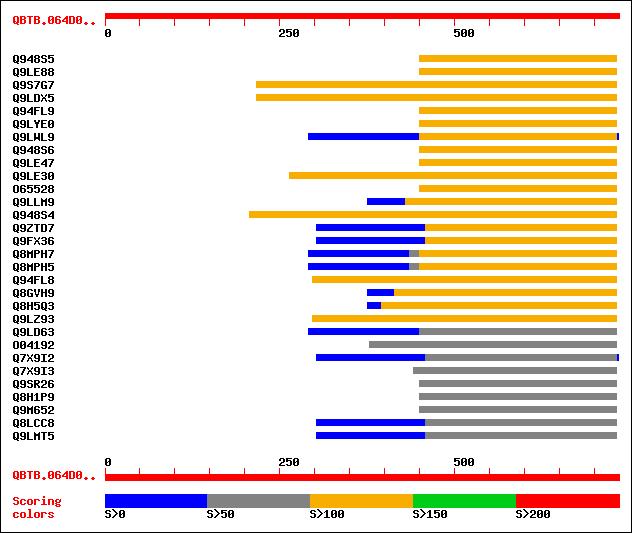
sptr|Q948S5|Q948S5 Myb. 121 9e-27
sptr|Q9LE88|Q9LE88 C-myb-like transcription factor (Fragment). 117 1e-25
sptr|Q9S7G7|Q9S7G7 Putative C-MYB-like transcription factor (Put... 116 3e-25
sptr|Q9LDX5|Q9LDX5 Putative c-myb-like transcription factor. 115 6e-25
sptr|Q94FL9|Q94FL9 Putative c-myb-like transcription factor MYB3... 115 8e-25
sptr|Q9LYE0|Q9LYE0 MYB like protein. 115 8e-25
sptr|Q9LWL9|Q9LWL9 Similar to Arabidopsis thaliana chromosome 4 ... 114 1e-24
sptr|Q948S6|Q948S6 Myb. 114 1e-24
sptr|Q9LE47|Q9LE47 Putative c-myb-like transcription factor (Fra... 114 2e-24
sptr|Q9LE30|Q9LE30 Putative c-myb-like transcription factor (Fra... 111 1e-23
sptr|O65528|O65528 Putative myb-protein (Fragment). 110 2e-23
sptr|Q9LLM9|Q9LLM9 Myb-like protein. 110 2e-23
sptr|Q948S4|Q948S4 Myb. 110 3e-23
sptr|Q9ZTD7|Q9ZTD7 Putative transcription factor. 103 2e-21
sptr|Q9FX36|Q9FX36 Myb-like transcription factor, putative. 103 2e-21
sptr|Q8MPH7|Q8MPH7 C-myb like protein. 103 3e-21
sptr|Q8MPH5|Q8MPH5 C-myb like protein. 103 3e-21
sptr|Q94FL8|Q94FL8 Putative c-myb-like transcription factor MYB3... 102 7e-21
sptr|Q8GVH9|Q8GVH9 P0592C06.14 protein. 102 7e-21
sptr|Q8H5Q3|Q8H5Q3 OJ1341_A08.36 protein. 102 7e-21
sptr|Q9LZ93|Q9LZ93 Myb-like protein. 102 7e-21
sptr|Q9LD63|Q9LD63 C-myb-like transcription factor (Fragment). 100 2e-20
sptr|O04192|O04192 MYB ISOLOG (Putative MYB family transcription... 100 2e-20
sptr|Q7X9I2|Q7X9I2 MYB transcription factor R2R3 type. 100 4e-20
sptr|Q7X9I3|Q7X9I3 MYB transcription factor R3 type. 100 4e-20
sptr|Q9SR26|Q9SR26 Putative MYB family transcription factor. 99 8e-20
sptr|Q8H1P9|Q8H1P9 Putative MYB family transcription factor. 99 8e-20
sptr|Q9M652|Q9M652 Putative c-myb-like transcription factor. 99 8e-20
sptr|Q8LCC8|Q8LCC8 Myb-like protein, putative. 98 1e-19
sptr|Q9LMT5|Q9LMT5 F2H15.17 protein. 98 1e-19
>sptr|Q948S5|Q948S5 Myb.
Length = 1042
Score = 121 bits (304), Expect = 9e-27
Identities = 51/94 (54%), Positives = 67/94 (71%)
Frame = +3
Query: 450 KHRWSSEEDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYLLDPAINKEPWSQQE 629
K WS EEDE + ++VK++G K W TIA +PGR C RW L+P INKE W+Q+E
Sbjct: 88 KGPWSKEEDEVIVELVKKYGPKKWSTIAQHLPGRIGKQCRERWHNHLNPGINKEAWTQEE 147
Query: 630 ELRLIRAQQVYGNKWCKMVKHFPGRTNDALKEHW 731
EL LIRA Q+YGNKW ++ K+ PGRT++A+K HW
Sbjct: 148 ELTLIRAHQIYGNKWAELTKYLPGRTDNAIKNHW 181
>sptr|Q9LE88|Q9LE88 C-myb-like transcription factor (Fragment).
Length = 98
Score = 117 bits (294), Expect = 1e-25
Identities = 49/94 (52%), Positives = 67/94 (71%)
Frame = +3
Query: 450 KHRWSSEEDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYLLDPAINKEPWSQQE 629
K WS EED+ + QMVK++G W TIA A+PGR C RW L+P INK+ W+Q+E
Sbjct: 5 KGPWSKEEDDVIIQMVKKYGPTKWSTIAQALPGRIGKQCRERWHNHLNPGINKDAWTQEE 64
Query: 630 ELRLIRAQQVYGNKWCKMVKHFPGRTNDALKEHW 731
E+RLI+A ++YGNKW ++ K PGRT++A+K HW
Sbjct: 65 EIRLIQAHRIYGNKWAELSKFLPGRTDNAIKNHW 98
>sptr|Q9S7G7|Q9S7G7 Putative C-MYB-like transcription factor
(Putative MYB-protein).
Length = 776
Score = 116 bits (291), Expect = 3e-25
Identities = 63/184 (34%), Positives = 93/184 (50%), Gaps = 12/184 (6%)
Frame = +3
Query: 216 PRADMEQSSDGLPISQRRANSSTKHSSKSDEVAEDMLQSNIPHETVGLCQGVESEEIEGP 395
P +E L Q R + + S+K P E LC+ VE +G
Sbjct: 8 PTTPLESLQGDLKGKQGRTSGPARRSTKGQWT---------PEEDEVLCKAVE--RFQGK 56
Query: 396 PHKKIVQNFTESS------------SSKNKKHRWSSEEDEALKQMVKRHGTKNWRTIACA 539
KKI + F + + + + K WS EED + +V+++G K W TI+
Sbjct: 57 NWKKIAECFKDRTDVQCLHRWQKVLNPELVKGPWSKEEDNTIIDLVEKYGPKKWSTISQH 116
Query: 540 IPGRNANSCLSRWKYLLDPAINKEPWSQQEELRLIRAQQVYGNKWCKMVKHFPGRTNDAL 719
+PGR C RW L+P INK W+Q+EEL LIRA Q+YGNKW +++K PGR+++++
Sbjct: 117 LPGRIGKQCRERWHNHLNPGINKNAWTQEEELTLIRAHQIYGNKWAELMKFLPGRSDNSI 176
Query: 720 KEHW 731
K HW
Sbjct: 177 KNHW 180
>sptr|Q9LDX5|Q9LDX5 Putative c-myb-like transcription factor.
Length = 776
Score = 115 bits (288), Expect = 6e-25
Identities = 64/185 (34%), Positives = 93/185 (50%), Gaps = 13/185 (7%)
Frame = +3
Query: 216 PRADMEQSSDGLPISQRRANSSTKHSSKSDEVAEDMLQSNIPHETVGLCQGVESEEIEGP 395
P +E L Q R + + S+K P E LC+ VE +G
Sbjct: 8 PTTPLESLQGDLKGKQGRTSGPARRSTKGQWT---------PEEDEVLCKAVE--RFQGK 56
Query: 396 PHKKIVQNFTESSSSKNKKHRW-------------SSEEDEALKQMVKRHGTKNWRTIAC 536
KKI + F + + + HRW S EED + +V+++G K W TI+
Sbjct: 57 NWKKIAECFKDRTDVQCL-HRWQKVLNPELVRGPWSKEEDNTIIDLVEKYGPKKWSTISQ 115
Query: 537 AIPGRNANSCLSRWKYLLDPAINKEPWSQQEELRLIRAQQVYGNKWCKMVKHFPGRTNDA 716
+PGR C RW L+P INK W+Q+EEL LIRA Q+YGNKW +++K PGR++++
Sbjct: 116 HLPGRIGKQCRERWHNHLNPGINKNAWTQEEELTLIRAHQIYGNKWAELMKFLPGRSDNS 175
Query: 717 LKEHW 731
+K HW
Sbjct: 176 IKNHW 180
>sptr|Q94FL9|Q94FL9 Putative c-myb-like transcription factor
MYB3R-4.
Length = 961
Score = 115 bits (287), Expect = 8e-25
Identities = 47/94 (50%), Positives = 66/94 (70%)
Frame = +3
Query: 450 KHRWSSEEDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYLLDPAINKEPWSQQE 629
K W+ EEDE + Q+++++G K W TIA +PGR C RW L+PAINKE W+Q+E
Sbjct: 81 KGPWTKEEDEMIVQLIEKYGPKKWSTIARFLPGRIGKQCRERWHNHLNPAINKEAWTQEE 140
Query: 630 ELRLIRAQQVYGNKWCKMVKHFPGRTNDALKEHW 731
EL LIRA Q+YGN+W ++ K PGR+++ +K HW
Sbjct: 141 ELLLIRAHQIYGNRWAELTKFLPGRSDNGIKNHW 174
>sptr|Q9LYE0|Q9LYE0 MYB like protein.
Length = 952
Score = 115 bits (287), Expect = 8e-25
Identities = 47/94 (50%), Positives = 66/94 (70%)
Frame = +3
Query: 450 KHRWSSEEDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYLLDPAINKEPWSQQE 629
K W+ EEDE + Q+++++G K W TIA +PGR C RW L+PAINKE W+Q+E
Sbjct: 81 KGPWTKEEDEMIVQLIEKYGPKKWSTIARFLPGRIGKQCRERWHNHLNPAINKEAWTQEE 140
Query: 630 ELRLIRAQQVYGNKWCKMVKHFPGRTNDALKEHW 731
EL LIRA Q+YGN+W ++ K PGR+++ +K HW
Sbjct: 141 ELLLIRAHQIYGNRWAELTKFLPGRSDNGIKNHW 174
>sptr|Q9LWL9|Q9LWL9 Similar to Arabidopsis thaliana chromosome 4
(Myb-like protein).
Length = 788
Score = 114 bits (285), Expect = 1e-24
Identities = 49/94 (52%), Positives = 64/94 (68%)
Frame = +3
Query: 450 KHRWSSEEDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYLLDPAINKEPWSQQE 629
K WS EEDE + QMV + G K W TIA A+PGR C RW L+P INKE W+Q+E
Sbjct: 102 KGPWSKEEDEIIVQMVNKLGPKKWSTIAQALPGRIGKQCRERWYNHLNPGINKEAWTQEE 161
Query: 630 ELRLIRAQQVYGNKWCKMVKHFPGRTNDALKEHW 731
E+ LI A ++YGNKW ++ K PGRT++++K HW
Sbjct: 162 EITLIHAHRMYGNKWAELTKFLPGRTDNSIKNHW 195
Score = 32.3 bits (72), Expect = 6.9
Identities = 23/105 (21%), Positives = 42/105 (40%)
Frame = +3
Query: 291 SSKSDEVAEDMLQSNIPHETVGLCQGVESEEIEGPPHKKIVQNFTESSSSKNKKHRWSSE 470
S + DE+ M+ P + + Q + G K+ + + + K W+ E
Sbjct: 106 SKEEDEIIVQMVNKLGPKKWSTIAQA-----LPGRIGKQCRERWYNHLNPGINKEAWTQE 160
Query: 471 EDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYLLDPAIN 605
E+ L + +G K W + +PGR NS + W + +N
Sbjct: 161 EEITLIHAHRMYGNK-WAELTKFLPGRTDNSIKNHWNSSVKKKVN 204
Score = 32.3 bits (72), Expect = 6.9
Identities = 16/44 (36%), Positives = 23/44 (52%), Gaps = 1/44 (2%)
Frame = +3
Query: 606 KEPWSQQEELRLIRAQQVY-GNKWCKMVKHFPGRTNDALKEHWK 734
K W+ +E+ L RA Q Y G W K+ + FP RT+ W+
Sbjct: 50 KGNWTPEEDAILSRAVQTYNGKNWKKIAECFPDRTDVQCLHRWQ 93
>sptr|Q948S6|Q948S6 Myb.
Length = 1003
Score = 114 bits (285), Expect = 1e-24
Identities = 50/94 (53%), Positives = 65/94 (69%)
Frame = +3
Query: 450 KHRWSSEEDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYLLDPAINKEPWSQQE 629
K W+ EED+ L ++V R+G K W TIA + GR C RW L+PAINKEPW+Q+E
Sbjct: 86 KGSWTKEEDDKLIELVNRYGPKKWSTIAQELAGRIGKQCRERWHNHLNPAINKEPWTQEE 145
Query: 630 ELRLIRAQQVYGNKWCKMVKHFPGRTNDALKEHW 731
EL LIRA QVYGNKW ++ K GR+++A+K HW
Sbjct: 146 ELTLIRAHQVYGNKWAELAKVLHGRSDNAIKNHW 179
>sptr|Q9LE47|Q9LE47 Putative c-myb-like transcription factor
(Fragment).
Length = 197
Score = 114 bits (284), Expect = 2e-24
Identities = 46/94 (48%), Positives = 66/94 (70%)
Frame = +3
Query: 450 KHRWSSEEDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYLLDPAINKEPWSQQE 629
K WS EED+ + +MVK++G K W TIA A+PGR C RW L+P INK+ W+Q+E
Sbjct: 25 KGPWSKEEDDIIVEMVKKYGPKKWSTIAQALPGRIGKQCRERWHNHLNPGINKDAWTQEE 84
Query: 630 ELRLIRAQQVYGNKWCKMVKHFPGRTNDALKEHW 731
E+ LI A ++YGNKW ++ K PG+T++++K HW
Sbjct: 85 EITLIHAHRMYGNKWAELTKFLPGKTDNSIKNHW 118
>sptr|Q9LE30|Q9LE30 Putative c-myb-like transcription factor
(Fragment).
Length = 599
Score = 111 bits (277), Expect = 1e-23
Identities = 60/168 (35%), Positives = 87/168 (51%), Gaps = 12/168 (7%)
Frame = +3
Query: 264 RRANSSTKHSSKSDEVAEDMLQSNIPHETVGLCQGVESEEIEGPPHKKIVQNFTESSSSK 443
RR T+ SSK P E L + V+ G KKI + FT+ + +
Sbjct: 21 RRTGGPTRRSSKGGWT---------PEEDETLRRAVQC--FNGKNWKKIAEFFTDRTDVQ 69
Query: 444 NK------------KHRWSSEEDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYL 587
K W+ EED+ + ++V ++G K W IA +PGR C RW
Sbjct: 70 CLHRWQKVLNPDLVKGAWTKEEDDRIMELVNKYGAKKWSVIAQNLPGRIGKQCRERWHNH 129
Query: 588 LDPAINKEPWSQQEELRLIRAQQVYGNKWCKMVKHFPGRTNDALKEHW 731
L+P+I +E W+QQE+L LIRA Q+YGNKW ++ K PGRT++++K HW
Sbjct: 130 LNPSIKREAWTQQEDLALIRAHQLYGNKWAEIAKFLPGRTDNSIKNHW 177
>sptr|O65528|O65528 Putative myb-protein (Fragment).
Length = 715
Score = 110 bits (275), Expect = 2e-23
Identities = 45/94 (47%), Positives = 64/94 (68%)
Frame = +3
Query: 450 KHRWSSEEDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYLLDPAINKEPWSQQE 629
K WS EED + +V+++G K W TI+ +PGR C RW L+P INK W+Q+E
Sbjct: 26 KGPWSKEEDNTIIDLVEKYGPKKWSTISQHLPGRIGKQCRERWHNHLNPGINKNAWTQEE 85
Query: 630 ELRLIRAQQVYGNKWCKMVKHFPGRTNDALKEHW 731
EL LIRA Q+YGNKW +++K PGR+++++K HW
Sbjct: 86 ELTLIRAHQIYGNKWAELMKFLPGRSDNSIKNHW 119
>sptr|Q9LLM9|Q9LLM9 Myb-like protein.
Length = 267
Score = 110 bits (275), Expect = 2e-23
Identities = 48/101 (47%), Positives = 70/101 (69%)
Frame = +3
Query: 429 SSSSKNKKHRWSSEEDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYLLDPAINK 608
SSSS + W EDE L+Q+V+++G +NW +IA + GR+ SC RW LDP INK
Sbjct: 19 SSSSACPRGHWRPGEDEKLRQLVEKYGPQNWNSIAEKLEGRSGKSCRLRWFNQLDPRINK 78
Query: 609 EPWSQQEELRLIRAQQVYGNKWCKMVKHFPGRTNDALKEHW 731
P++++EE RL+ A + +GNKW + +HFPGRT++A+K HW
Sbjct: 79 RPFTEEEEERLLAAHRHHGNKWALIARHFPGRTDNAVKNHW 119
Score = 35.4 bits (80), Expect = 0.82
Identities = 20/68 (29%), Positives = 31/68 (45%)
Frame = +3
Query: 375 SEEIEGPPHKKIVQNFTESSSSKNKKHRWSSEEDEALKQMVKRHGTKNWRTIACAIPGRN 554
+E++EG K + + K ++ EE+E L + HG K W IA PGR
Sbjct: 53 AEKLEGRSGKSCRLRWFNQLDPRINKRPFTEEEEERLLAAHRHHGNK-WALIARHFPGRT 111
Query: 555 ANSCLSRW 578
N+ + W
Sbjct: 112 DNAVKNHW 119
>sptr|Q948S4|Q948S4 Myb.
Length = 588
Score = 110 bits (274), Expect = 3e-23
Identities = 61/179 (34%), Positives = 96/179 (53%), Gaps = 4/179 (2%)
Frame = +3
Query: 207 ASYPRADMEQSSDGLPISQRRANSSTKHSSKSDEVAEDMLQSNIPHETVGLCQGVESEEI 386
AS PR E S S RR++ TK SS++ E+ N+ E V +G ++I
Sbjct: 29 ASTPRYSSEPGS-----SYRRSSGPTKRSSQAGWTEEE---DNLLTEVVKRFKGRNWKKI 80
Query: 387 ----EGPPHKKIVQNFTESSSSKNKKHRWSSEEDEALKQMVKRHGTKNWRTIACAIPGRN 554
G + + + + + + K WS EED+ + ++V+++G K W IA ++PGR
Sbjct: 81 AECMNGRTDVQCLHRWQKVLNPELVKGPWSKEEDDLIVELVEKYGCKKWSFIAKSMPGRI 140
Query: 555 ANSCLSRWKYLLDPAINKEPWSQQEELRLIRAQQVYGNKWCKMVKHFPGRTNDALKEHW 731
C RW LDP I ++ W++QEE L Q+YGNKW ++ + PGRT++A+K HW
Sbjct: 141 GKQCRERWHNHLDPTIKRDAWTEQEESVLCHYHQIYGNKWAEIARFLPGRTDNAIKNHW 199
>sptr|Q9ZTD7|Q9ZTD7 Putative transcription factor.
Length = 243
Score = 103 bits (257), Expect = 2e-21
Identities = 42/91 (46%), Positives = 63/91 (69%)
Frame = +3
Query: 459 WSSEEDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYLLDPAINKEPWSQQEELR 638
W EDE LK +V+++G NW IA +PGR+ SC RW LDP IN+ P++++EE R
Sbjct: 9 WRPAEDEKLKDLVEQYGPHNWNAIALKLPGRSGKSCRLRWFNQLDPRINRNPFTEEEEER 68
Query: 639 LIRAQQVYGNKWCKMVKHFPGRTNDALKEHW 731
L+ A +++GN+W + + FPGRT++A+K HW
Sbjct: 69 LLAAHRIHGNRWSIIARLFPGRTDNAVKNHW 99
Score = 32.0 bits (71), Expect = 9.0
Identities = 21/96 (21%), Positives = 41/96 (42%)
Frame = +3
Query: 303 DEVAEDMLQSNIPHETVGLCQGVESEEIEGPPHKKIVQNFTESSSSKNKKHRWSSEEDEA 482
DE +D+++ PH + ++ G K + + ++ ++ EE+E
Sbjct: 14 DEKLKDLVEQYGPHNWNAIAL-----KLPGRSGKSCRLRWFNQLDPRINRNPFTEEEEER 68
Query: 483 LKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYLL 590
L + HG + W IA PGR N+ + W ++
Sbjct: 69 LLAAHRIHGNR-WSIIARLFPGRTDNAVKNHWHVIM 103
>sptr|Q9FX36|Q9FX36 Myb-like transcription factor, putative.
Length = 243
Score = 103 bits (257), Expect = 2e-21
Identities = 42/91 (46%), Positives = 63/91 (69%)
Frame = +3
Query: 459 WSSEEDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYLLDPAINKEPWSQQEELR 638
W EDE LK +V+++G NW IA +PGR+ SC RW LDP IN+ P++++EE R
Sbjct: 9 WRPAEDEKLKDLVEQYGPHNWNAIALKLPGRSGKSCRLRWFNQLDPRINRNPFTEEEEER 68
Query: 639 LIRAQQVYGNKWCKMVKHFPGRTNDALKEHW 731
L+ A +++GN+W + + FPGRT++A+K HW
Sbjct: 69 LLAAHRIHGNRWSIIARLFPGRTDNAVKNHW 99
Score = 32.0 bits (71), Expect = 9.0
Identities = 21/96 (21%), Positives = 41/96 (42%)
Frame = +3
Query: 303 DEVAEDMLQSNIPHETVGLCQGVESEEIEGPPHKKIVQNFTESSSSKNKKHRWSSEEDEA 482
DE +D+++ PH + ++ G K + + ++ ++ EE+E
Sbjct: 14 DEKLKDLVEQYGPHNWNAIAL-----KLPGRSGKSCRLRWFNQLDPRINRNPFTEEEEER 68
Query: 483 LKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYLL 590
L + HG + W IA PGR N+ + W ++
Sbjct: 69 LLAAHRIHGNR-WSIIARLFPGRTDNAVKNHWHVIM 103
>sptr|Q8MPH7|Q8MPH7 C-myb like protein.
Length = 436
Score = 103 bits (256), Expect = 3e-21
Identities = 44/94 (46%), Positives = 61/94 (64%)
Frame = +3
Query: 450 KHRWSSEEDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYLLDPAINKEPWSQQE 629
K W+ EEDE L+QMV+ HG +NW TIA A+PGR C RW LDP I KE WS +E
Sbjct: 145 KGPWTKEEDEILRQMVETHGPRNWSTIAAALPGRIGKQCRERWHNHLDPNIKKEKWSPEE 204
Query: 630 ELRLIRAQQVYGNKWCKMVKHFPGRTNDALKEHW 731
+ +++ GN+WC++ K PGRT++A+K +
Sbjct: 205 DRIILKMHLEMGNRWCEIAKLLPGRTDNAIKNRF 238
Score = 56.2 bits (134), Expect = 4e-07
Identities = 28/101 (27%), Positives = 51/101 (50%), Gaps = 2/101 (1%)
Frame = +3
Query: 435 SSKNKKHRWSSEEDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYLLDPA-INKE 611
+S+ KK W+ +D L + V++ G + W I+ + R+ C +RW L D ++K
Sbjct: 87 ASQKKKVEWTPFDDAKLLKYVEKFGDQKWNKISKFMRDRSEIQCFNRWLELKDTCFVSKG 146
Query: 612 PWSQQEELRLIRAQQVYG-NKWCKMVKHFPGRTNDALKEHW 731
PW+++E+ L + + +G W + PGR +E W
Sbjct: 147 PWTKEEDEILRQMVETHGPRNWSTIAAALPGRIGKQCRERW 187
Score = 38.1 bits (87), Expect = 0.13
Identities = 27/121 (22%), Positives = 57/121 (47%), Gaps = 1/121 (0%)
Frame = +3
Query: 291 SSKSDEVAEDMLQSNIPHETVGLCQGVESEEIEGPPHKKIVQNFTESSSSKNKKHRWSSE 470
+ + DE+ M++++ P + + G K+ + + KK +WS E
Sbjct: 149 TKEEDEILRQMVETHGPRNWSTIAAA-----LPGRIGKQCRERWHNHLDPNIKKEKWSPE 203
Query: 471 EDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYLLDPAINKEP-WSQQEELRLIR 647
ED + +M G + W IA +PGR N+ +R+ L I+++ ++ ++L +I+
Sbjct: 204 EDRIILKMHLEMGNR-WCEIAKLLPGRTDNAIKNRFNSKLKKQISRQSNFAASQQLDIIQ 262
Query: 648 A 650
+
Sbjct: 263 S 263
>sptr|Q8MPH5|Q8MPH5 C-myb like protein.
Length = 436
Score = 103 bits (256), Expect = 3e-21
Identities = 44/94 (46%), Positives = 61/94 (64%)
Frame = +3
Query: 450 KHRWSSEEDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYLLDPAINKEPWSQQE 629
K W+ EEDE L+QMV+ HG +NW TIA A+PGR C RW LDP I KE WS +E
Sbjct: 145 KGPWTKEEDEILRQMVETHGPRNWSTIAAALPGRIGKQCRERWHNHLDPNIKKEKWSPEE 204
Query: 630 ELRLIRAQQVYGNKWCKMVKHFPGRTNDALKEHW 731
+ +++ GN+WC++ K PGRT++A+K +
Sbjct: 205 DRIILKMHLEMGNRWCEIAKLLPGRTDNAIKNRF 238
Score = 56.2 bits (134), Expect = 4e-07
Identities = 28/101 (27%), Positives = 51/101 (50%), Gaps = 2/101 (1%)
Frame = +3
Query: 435 SSKNKKHRWSSEEDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYLLDPA-INKE 611
+S+ KK W+ +D L + V++ G + W I+ + R+ C +RW L D ++K
Sbjct: 87 ASQKKKVEWTPFDDAKLLKYVEKFGDQKWNKISKFMRDRSEIQCFNRWLELKDTCFVSKG 146
Query: 612 PWSQQEELRLIRAQQVYG-NKWCKMVKHFPGRTNDALKEHW 731
PW+++E+ L + + +G W + PGR +E W
Sbjct: 147 PWTKEEDEILRQMVETHGPRNWSTIAAALPGRIGKQCRERW 187
Score = 38.1 bits (87), Expect = 0.13
Identities = 27/121 (22%), Positives = 57/121 (47%), Gaps = 1/121 (0%)
Frame = +3
Query: 291 SSKSDEVAEDMLQSNIPHETVGLCQGVESEEIEGPPHKKIVQNFTESSSSKNKKHRWSSE 470
+ + DE+ M++++ P + + G K+ + + KK +WS E
Sbjct: 149 TKEEDEILRQMVETHGPRNWSTIAAA-----LPGRIGKQCRERWHNHLDPNIKKEKWSPE 203
Query: 471 EDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYLLDPAINKEP-WSQQEELRLIR 647
ED + +M G + W IA +PGR N+ +R+ L I+++ ++ ++L +I+
Sbjct: 204 EDRIILKMHLEMGNR-WCEIAKLLPGRTDNAIKNRFNSKLKKQISRQSNFAASQQLDIIQ 262
Query: 648 A 650
+
Sbjct: 263 S 263
>sptr|Q94FL8|Q94FL8 Putative c-myb-like transcription factor
MYB3R-5.
Length = 548
Score = 102 bits (253), Expect = 7e-21
Identities = 54/171 (31%), Positives = 86/171 (50%), Gaps = 26/171 (15%)
Frame = +3
Query: 297 KSDEVAEDMLQSNIPHETVGLCQGVES--------------EEIEGPPHKKIVQNFTESS 434
KS E+A S+ P T G + + E+ +G KKI + F E +
Sbjct: 50 KSPEIATPATVSSFPRRTSGPMRRAKGGWTPEEDETLRRAVEKYKGKRWKKIAEFFPERT 109
Query: 435 ------------SSKNKKHRWSSEEDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSRW 578
+ + K W+ EED+ + ++VK++G W IA ++PGR C RW
Sbjct: 110 EVQCLHRWQKVLNPELVKGPWTQEEDDKIVELVKKYGPAKWSVIAKSLPGRIGKQCRERW 169
Query: 579 KYLLDPAINKEPWSQQEELRLIRAQQVYGNKWCKMVKHFPGRTNDALKEHW 731
L+P I K+ W+ +EE L+ + ++YGNKW ++ K PGRT++A+K HW
Sbjct: 170 HNHLNPGIRKDAWTVEEESALMNSHRMYGNKWAEIAKVLPGRTDNAIKNHW 220
>sptr|Q8GVH9|Q8GVH9 P0592C06.14 protein.
Length = 438
Score = 102 bits (253), Expect = 7e-21
Identities = 45/110 (40%), Positives = 64/110 (58%), Gaps = 4/110 (3%)
Frame = +3
Query: 414 QNFTESSSSKNK----KHRWSSEEDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSRWK 581
Q T+++ S + K W+ EEDE L+QMV+ HG + W IA ++PGR C RW
Sbjct: 92 QQQTQAAGSNDNPGLVKGGWTREEDEVLRQMVRHHGDRKWAEIAKSLPGRVGKQCRERWT 151
Query: 582 YLLDPAINKEPWSQQEELRLIRAQQVYGNKWCKMVKHFPGRTNDALKEHW 731
L P I K+ W+++E+ LI A Q YGN W + K PGR+ + +K HW
Sbjct: 152 NHLHPDIKKDIWTEEEDRMLIEAHQTYGNSWSAIAKQLPGRSENTIKNHW 201
Score = 36.6 bits (83), Expect = 0.37
Identities = 19/68 (27%), Positives = 34/68 (50%)
Frame = +3
Query: 375 SEEIEGPPHKKIVQNFTESSSSKNKKHRWSSEEDEALKQMVKRHGTKNWRTIACAIPGRN 554
++ + G K+ + +T KK W+ EED L + + +G +W IA +PGR+
Sbjct: 135 AKSLPGRVGKQCRERWTNHLHPDIKKDIWTEEEDRMLIEAHQTYG-NSWSAIAKQLPGRS 193
Query: 555 ANSCLSRW 578
N+ + W
Sbjct: 194 ENTIKNHW 201
>sptr|Q8H5Q3|Q8H5Q3 OJ1341_A08.36 protein.
Length = 329
Score = 102 bits (253), Expect = 7e-21
Identities = 44/112 (39%), Positives = 68/112 (60%)
Frame = +3
Query: 396 PHKKIVQNFTESSSSKNKKHRWSSEEDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSR 575
P ++ +Q + +S+S K W+ EEDE L+QMV+ HG + W IA ++PGR C R
Sbjct: 76 PQQQQLQAASSNSNSGLIKGGWTREEDEVLRQMVRHHGDRKWAEIAKSLPGRIGKQCRER 135
Query: 576 WKYLLDPAINKEPWSQQEELRLIRAQQVYGNKWCKMVKHFPGRTNDALKEHW 731
W L P I K W+++E+ +LI+A Q YGN+W + + PGR+ + +K W
Sbjct: 136 WTNHLHPDIKKGIWTEEEDRKLIKAHQTYGNRWSAIARSLPGRSENTVKNRW 187
Score = 38.9 bits (89), Expect = 0.074
Identities = 21/70 (30%), Positives = 37/70 (52%), Gaps = 2/70 (2%)
Frame = +3
Query: 375 SEEIEGPPHKKIVQNFTESSSSKNKKHRWSSEEDEALKQMVKRHGT--KNWRTIACAIPG 548
++ + G K+ + +T KK W+ EED ++++K H T W IA ++PG
Sbjct: 121 AKSLPGRIGKQCRERWTNHLHPDIKKGIWTEEED---RKLIKAHQTYGNRWSAIARSLPG 177
Query: 549 RNANSCLSRW 578
R+ N+ +RW
Sbjct: 178 RSENTVKNRW 187
>sptr|Q9LZ93|Q9LZ93 Myb-like protein.
Length = 529
Score = 102 bits (253), Expect = 7e-21
Identities = 54/171 (31%), Positives = 86/171 (50%), Gaps = 26/171 (15%)
Frame = +3
Query: 297 KSDEVAEDMLQSNIPHETVGLCQGVES--------------EEIEGPPHKKIVQNFTESS 434
KS E+A S+ P T G + + E+ +G KKI + F E +
Sbjct: 31 KSPEIATPATVSSFPRRTSGPMRRAKGGWTPEEDETLRRAVEKYKGKRWKKIAEFFPERT 90
Query: 435 ------------SSKNKKHRWSSEEDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSRW 578
+ + K W+ EED+ + ++VK++G W IA ++PGR C RW
Sbjct: 91 EVQCLHRWQKVLNPELVKGPWTQEEDDKIVELVKKYGPAKWSVIAKSLPGRIGKQCRERW 150
Query: 579 KYLLDPAINKEPWSQQEELRLIRAQQVYGNKWCKMVKHFPGRTNDALKEHW 731
L+P I K+ W+ +EE L+ + ++YGNKW ++ K PGRT++A+K HW
Sbjct: 151 HNHLNPGIRKDAWTVEEESALMNSHRMYGNKWAEIAKVLPGRTDNAIKNHW 201
>sptr|Q9LD63|Q9LD63 C-myb-like transcription factor (Fragment).
Length = 254
Score = 100 bits (250), Expect = 2e-20
Identities = 42/94 (44%), Positives = 59/94 (62%)
Frame = +3
Query: 450 KHRWSSEEDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYLLDPAINKEPWSQQE 629
K W+ EEDE + ++V ++G K W +A ++PGR C RW LDP I K+ W+ +E
Sbjct: 25 KGPWTKEEDEKIAELVNKNGPKKWSVVARSLPGRIGKQCRERWHNHLDPHIKKDAWTPEE 84
Query: 630 ELRLIRAQQVYGNKWCKMVKHFPGRTNDALKEHW 731
E LI A Q GNKW ++ K PGRT++A+K HW
Sbjct: 85 EQALIEAHQRNGNKWAEIAKSLPGRTDNAIKNHW 118
Score = 40.8 bits (94), Expect = 0.019
Identities = 25/96 (26%), Positives = 46/96 (47%)
Frame = +3
Query: 291 SSKSDEVAEDMLQSNIPHETVGLCQGVESEEIEGPPHKKIVQNFTESSSSKNKKHRWSSE 470
+ + DE +++ N P + V + + G K+ + + KK W+ E
Sbjct: 29 TKEEDEKIAELVNKNGPKKW-----SVVARSLPGRIGKQCRERWHNHLDPHIKKDAWTPE 83
Query: 471 EDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSRW 578
E++AL + +R+G K W IA ++PGR N+ + W
Sbjct: 84 EEQALIEAHQRNGNK-WAEIAKSLPGRTDNAIKNHW 118
>sptr|O04192|O04192 MYB ISOLOG (Putative MYB family transcription
factor).
Length = 367
Score = 100 bits (249), Expect = 2e-20
Identities = 51/125 (40%), Positives = 72/125 (57%), Gaps = 7/125 (5%)
Frame = +3
Query: 378 EEIEGPPHKKIVQNFTESSSS------KNK-KHRWSSEEDEALKQMVKRHGTKNWRTIAC 536
+ E HK + E + S K+K K W E+DEAL ++VK G +NW I+
Sbjct: 19 KSFENAIHKAVEAELAELAKSDANGGGKSKVKGPWLPEQDEALTRLVKMCGPRNWNLISR 78
Query: 537 AIPGRNANSCLSRWKYLLDPAINKEPWSQQEELRLIRAQQVYGNKWCKMVKHFPGRTNDA 716
IPGR+ SC RW LDP + ++P+S +EE ++ AQ V GNKW + K PGRT++A
Sbjct: 79 GIPGRSGKSCRLRWCNQLDPILKRKPFSDEEEHMIMSAQAVLGNKWSVIAKLLPGRTDNA 138
Query: 717 LKEHW 731
+K HW
Sbjct: 139 IKNHW 143
>sptr|Q7X9I2|Q7X9I2 MYB transcription factor R2R3 type.
Length = 182
Score = 99.8 bits (247), Expect = 4e-20
Identities = 40/91 (43%), Positives = 63/91 (69%)
Frame = +3
Query: 459 WSSEEDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYLLDPAINKEPWSQQEELR 638
W EDE LK++V+++G NW IA + GR+ SC RW LDP IN+ P++++EE R
Sbjct: 7 WRPAEDEKLKELVEKYGPHNWNAIAEKLHGRSGKSCRLRWFNQLDPRINRSPFTEEEEER 66
Query: 639 LIRAQQVYGNKWCKMVKHFPGRTNDALKEHW 731
L+ + +++GN+W + + FPGRT++A+K HW
Sbjct: 67 LLASHRIHGNRWAVIARLFPGRTDNAVKNHW 97
Score = 40.8 bits (94), Expect = 0.019
Identities = 31/163 (19%), Positives = 68/163 (41%), Gaps = 19/163 (11%)
Frame = +3
Query: 303 DEVAEDMLQSNIPHETVGLCQGVESEEIEGPPHKKIVQNFTESSSSKNKKHRWSSEEDEA 482
DE +++++ PH + +E++ G K + + + ++ EE+E
Sbjct: 12 DEKLKELVEKYGPHNWNAI-----AEKLHGRSGKSCRLRWFNQLDPRINRSPFTEEEEER 66
Query: 483 LKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYLL----------------DPAINKEP 614
L + HG + W IA PGR N+ + W ++ +N++
Sbjct: 67 LLASHRIHGNR-WAVIARLFPGRTDNAVKNHWHVIMARRYRERSRLHAKRTAQALVNEQN 125
Query: 615 WSQQEELRL---IRAQQVYGNKWCKMVKHFPGRTNDALKEHWK 734
+S ++++++ R+ + K+C+ +P T+ L WK
Sbjct: 126 FSSKQDMQINCETRSFSSFVKKYCEKFGQYPLITHSYLPAFWK 168
>sptr|Q7X9I3|Q7X9I3 MYB transcription factor R3 type.
Length = 530
Score = 99.8 bits (247), Expect = 4e-20
Identities = 40/94 (42%), Positives = 61/94 (64%)
Frame = +3
Query: 450 KHRWSSEEDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYLLDPAINKEPWSQQE 629
K W+ EED+ + ++V ++G W IA A+PGR C RW L+P I K+ W+ +E
Sbjct: 112 KGPWTQEEDDKIVELVAKYGPTKWSVIAKALPGRIGKQCRERWHNHLNPDIKKDAWTLEE 171
Query: 630 ELRLIRAQQVYGNKWCKMVKHFPGRTNDALKEHW 731
EL L+ A ++YGNKW ++ K PGRT++++K HW
Sbjct: 172 ELALMNAHRIYGNKWAEIAKVLPGRTDNSIKNHW 205
Score = 78.6 bits (192), Expect = 8e-14
Identities = 36/98 (36%), Positives = 54/98 (55%), Gaps = 1/98 (1%)
Frame = +3
Query: 441 KNKKHRWSSEEDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYLLDPAINKEPWS 620
+ K W+ EEDE L+ V + K+W+ IA P R+ CL RW+ +LDP + K PW+
Sbjct: 57 RRAKGGWTPEEDETLRTAVATYRGKSWKKIAEFFPDRSEVQCLHRWQKVLDPELVKGPWT 116
Query: 621 QQEELRLIRAQQVYG-NKWCKMVKHFPGRTNDALKEHW 731
Q+E+ +++ YG KW + K PGR +E W
Sbjct: 117 QEEDDKIVELVAKYGPTKWSVIAKALPGRIGKQCRERW 154
>sptr|Q9SR26|Q9SR26 Putative MYB family transcription factor.
Length = 496
Score = 98.6 bits (244), Expect = 8e-20
Identities = 40/94 (42%), Positives = 62/94 (65%)
Frame = +3
Query: 450 KHRWSSEEDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYLLDPAINKEPWSQQE 629
K W+ EEDE + ++V+++G W IA ++PGR C RW L+P INK+ W+ +E
Sbjct: 121 KGPWTHEEDEKIVELVEKYGPAKWSIIAQSLPGRIGKQCRERWHNHLNPDINKDAWTTEE 180
Query: 630 ELRLIRAQQVYGNKWCKMVKHFPGRTNDALKEHW 731
E+ L+ A + +GNKW ++ K PGRT++A+K HW
Sbjct: 181 EVALMNAHRSHGNKWAEIAKVLPGRTDNAIKNHW 214
>sptr|Q8H1P9|Q8H1P9 Putative MYB family transcription factor.
Length = 505
Score = 98.6 bits (244), Expect = 8e-20
Identities = 40/94 (42%), Positives = 62/94 (65%)
Frame = +3
Query: 450 KHRWSSEEDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYLLDPAINKEPWSQQE 629
K W+ EEDE + ++V+++G W IA ++PGR C RW L+P INK+ W+ +E
Sbjct: 130 KGPWTHEEDEKIVELVEKYGPAKWSIIAQSLPGRIGKQCRERWHNHLNPDINKDAWTTEE 189
Query: 630 ELRLIRAQQVYGNKWCKMVKHFPGRTNDALKEHW 731
E+ L+ A + +GNKW ++ K PGRT++A+K HW
Sbjct: 190 EVALMNAHRSHGNKWAEIAKVLPGRTDNAIKNHW 223
>sptr|Q9M652|Q9M652 Putative c-myb-like transcription factor.
Length = 505
Score = 98.6 bits (244), Expect = 8e-20
Identities = 40/94 (42%), Positives = 62/94 (65%)
Frame = +3
Query: 450 KHRWSSEEDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYLLDPAINKEPWSQQE 629
K W+ EEDE + ++V+++G W IA ++PGR C RW L+P INK+ W+ +E
Sbjct: 130 KGPWTHEEDEKIVELVEKYGPAKWSIIAQSLPGRIGKQCRERWHNHLNPDINKDAWTTEE 189
Query: 630 ELRLIRAQQVYGNKWCKMVKHFPGRTNDALKEHW 731
E+ L+ A + +GNKW ++ K PGRT++A+K HW
Sbjct: 190 EVALMNAHRSHGNKWAEIAKVLPGRTDNAIKNHW 223
>sptr|Q8LCC8|Q8LCC8 Myb-like protein, putative.
Length = 249
Score = 98.2 bits (243), Expect = 1e-19
Identities = 39/91 (42%), Positives = 62/91 (68%)
Frame = +3
Query: 459 WSSEEDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYLLDPAINKEPWSQQEELR 638
W EDE L+++V++ G NW IA + GR+ SC RW LDP IN+ P++++EE R
Sbjct: 8 WRPAEDEKLRELVEQFGPHNWNAIAQKLSGRSGKSCRLRWFNQLDPRINRNPFTEEEEER 67
Query: 639 LIRAQQVYGNKWCKMVKHFPGRTNDALKEHW 731
L+ + +++GN+W + + FPGRT++A+K HW
Sbjct: 68 LLASHRIHGNRWSVIARFFPGRTDNAVKNHW 98
Score = 32.7 bits (73), Expect = 5.3
Identities = 21/96 (21%), Positives = 41/96 (42%)
Frame = +3
Query: 303 DEVAEDMLQSNIPHETVGLCQGVESEEIEGPPHKKIVQNFTESSSSKNKKHRWSSEEDEA 482
DE ++++ PH + Q ++ G K + + ++ ++ EE+E
Sbjct: 13 DEKLRELVEQFGPHNWNAIAQ-----KLSGRSGKSCRLRWFNQLDPRINRNPFTEEEEER 67
Query: 483 LKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYLL 590
L + HG + W IA PGR N+ + W ++
Sbjct: 68 LLASHRIHGNR-WSVIARFFPGRTDNAVKNHWHVIM 102
>sptr|Q9LMT5|Q9LMT5 F2H15.17 protein.
Length = 248
Score = 98.2 bits (243), Expect = 1e-19
Identities = 39/91 (42%), Positives = 62/91 (68%)
Frame = +3
Query: 459 WSSEEDEALKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYLLDPAINKEPWSQQEELR 638
W EDE L+++V++ G NW IA + GR+ SC RW LDP IN+ P++++EE R
Sbjct: 7 WRPAEDEKLRELVEQFGPHNWNAIAQKLSGRSGKSCRLRWFNQLDPRINRNPFTEEEEER 66
Query: 639 LIRAQQVYGNKWCKMVKHFPGRTNDALKEHW 731
L+ + +++GN+W + + FPGRT++A+K HW
Sbjct: 67 LLASHRIHGNRWSVIARFFPGRTDNAVKNHW 97
Score = 32.7 bits (73), Expect = 5.3
Identities = 21/96 (21%), Positives = 41/96 (42%)
Frame = +3
Query: 303 DEVAEDMLQSNIPHETVGLCQGVESEEIEGPPHKKIVQNFTESSSSKNKKHRWSSEEDEA 482
DE ++++ PH + Q ++ G K + + ++ ++ EE+E
Sbjct: 12 DEKLRELVEQFGPHNWNAIAQ-----KLSGRSGKSCRLRWFNQLDPRINRNPFTEEEEER 66
Query: 483 LKQMVKRHGTKNWRTIACAIPGRNANSCLSRWKYLL 590
L + HG + W IA PGR N+ + W ++
Sbjct: 67 LLASHRIHGNR-WSVIARFFPGRTDNAVKNHWHVIM 101
Database: swall
Posted date: Feb 26, 2004 3:00 PM
Number of letters in database: 439,479,560
Number of sequences in database: 1,381,838
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 592,268,853
Number of Sequences: 1381838
Number of extensions: 12287476
Number of successful extensions: 38115
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 35516
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 37290
length of database: 439,479,560
effective HSP length: 120
effective length of database: 273,659,000
effective search space used: 33933716000
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

