BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
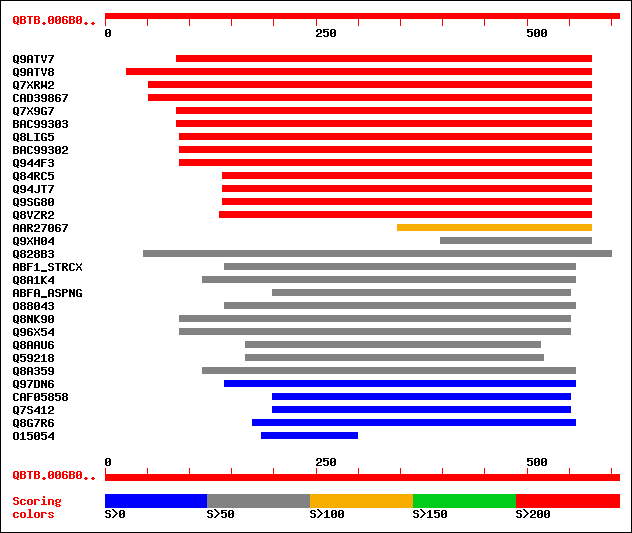
Query= QBTB.006B01F020311.3.1
(609 letters)
Database: swall
1,381,838 sequences; 439,479,560 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sptr|Q9ATV7|Q9ATV7 Arabinoxylan arabinofuranohydrolase isoenzyme... 263 1e-69
sptr|Q9ATV8|Q9ATV8 Arabinoxylan arabinofuranohydrolase isoenzyme... 261 5e-69
sptr|Q7XRW2|Q7XRW2 OSJNBb0058J09.6 protein. 258 3e-68
sptrnew|CAD39867|CAD39867 OSJNBb0058J09.4 protein. 258 3e-68
sptr|Q7X9G7|Q7X9G7 Alpha-L-arabinofuranosidase. 225 3e-58
sptrnew|BAC99303|BAC99303 Alpha-L-arabinofuranosidase. 223 1e-57
sptr|Q8LIG5|Q8LIG5 Putative arabinoxylan narabinofuranohydrolase... 219 2e-56
sptrnew|BAC99302|BAC99302 Alpha-L-arabinofuranosidase. 215 3e-55
sptr|Q944F3|Q944F3 Arabinosidase ARA-1. 215 3e-55
sptr|Q84RC5|Q84RC5 Alpha-L-arabinofuranosidase. 208 4e-53
sptr|Q94JT7|Q94JT7 AT3g10740/T7M13_18. 208 4e-53
sptr|Q9SG80|Q9SG80 Putative alpha-L-arabinofuranosidase. 208 4e-53
sptr|Q8VZR2|Q8VZR2 Putative arabinosidase (Alpha-L-arabinofurano... 204 7e-52
sptrnew|AAR27067|AAR27067 A-arabinofuranosidase 1 (Fragment). 102 4e-21
sptr|Q9XH04|Q9XH04 T1N24.13 protein. 81 1e-14
sptr|Q828B3|Q828B3 Putative secreted arabinosidase. 60 2e-08
sw|P82593|ABF1_STRCX Alpha-L-arabinofuranosidase I precursor (EC... 59 4e-08
sptr|Q8A1K4|Q8A1K4 Alpha-L-arabinofuranosidase A precursor. 59 5e-08
sw|P42254|ABFA_ASPNG Alpha-L-arabinofuranosidase A precursor (EC... 59 6e-08
sptr|O88043|O88043 Putative secreted arabinosidase. 58 1e-07
sptr|Q8NK90|Q8NK90 Alpha-L-arabinofuranosidase A (EC 3.2.1.55). 57 2e-07
sptr|Q96X54|Q96X54 Alpha-L-arabinofuranosidase A (EC 3.2.1.55). 57 2e-07
sptr|Q8AAU6|Q8AAU6 Alpha-L-arabinofuranosidase A precursor. 53 3e-06
sptr|Q59218|Q59218 Arabinosidase (EC 3.2.1.55). 51 1e-05
sptr|Q8A359|Q8A359 Alpha-L-arabinofuranosidase A precursor. 51 1e-05
sptr|Q97DN6|Q97DN6 Probable alpha-arabinofuranosidase. 49 5e-05
sptrnew|CAF05858|CAF05858 Related to alpha-L-arabinofuranosidase A. 47 2e-04
sptr|Q7S412|Q7S412 Hypothetical protein. 47 2e-04
sptr|Q8G7R6|Q8G7R6 Similar to alpha-arabinofuranosidase I. 43 0.004
sptr|O15054|O15054 Hypothetical protein KIAA0346 (Fragment). 37 0.20
>sptr|Q9ATV7|Q9ATV7 Arabinoxylan arabinofuranohydrolase isoenzyme
AXAH-II.
Length = 656
Score = 263 bits (672), Expect = 1e-69
Identities = 127/164 (77%), Positives = 141/164 (85%)
Frame = +1
Query: 85 CIASKCLGGASGLNSTQMVTLKVDASPKLARKIPDTFLGVFFEEMGHGGAGGIWAELVSN 264
C KC+ AS L +TQ TLKVDAS +LARKIPDT G+FFEE+ H GAGGIWAELVSN
Sbjct: 19 CFGCKCI--ASELQATQTATLKVDASSQLARKIPDTLFGMFFEEINHAGAGGIWAELVSN 76
Query: 265 RGFEAGGPNTPSNIDPWLIVGDDSSVYVETDRSSCFSRNIVALRMEVLCNDCPAGGVGIY 444
RGFEAGG +TPSNIDPW I+GDDSS++V TDR+SCFSRNI+ALRMEVLC+DCPA GVGIY
Sbjct: 77 RGFEAGGLHTPSNIDPWSIIGDDSSIFVATDRTSCFSRNIIALRMEVLCDDCPASGVGIY 136
Query: 445 NPGFWGMNIEDGKTYHLVMYVKSPKTTCLTVSLTSSDGLQNLAS 576
NPGFWGMNIEDGKTY+LVMYVKS + LTVSL SSDGLQNLAS
Sbjct: 137 NPGFWGMNIEDGKTYNLVMYVKSAEAAELTVSLASSDGLQNLAS 180
>sptr|Q9ATV8|Q9ATV8 Arabinoxylan arabinofuranohydrolase isoenzyme
AXAH-I.
Length = 658
Score = 261 bits (667), Expect = 5e-69
Identities = 129/184 (70%), Positives = 150/184 (81%)
Frame = +1
Query: 25 GSNSMEGPPRTLIHXXXXXXCIASKCLGGASGLNSTQMVTLKVDASPKLARKIPDTFLGV 204
GS M RT++ C++ KCL AS TQ+ +L VD+SP LARKIPDT G+
Sbjct: 2 GSKEMPRRTRTIL-CFLLLFCVSCKCL--ASEFEITQVASLGVDSSPHLARKIPDTLFGI 58
Query: 205 FFEEMGHGGAGGIWAELVSNRGFEAGGPNTPSNIDPWLIVGDDSSVYVETDRSSCFSRNI 384
FFEE+ H GAGGIWAELVSNRGFEAGGP+TPSNI+PW I+GDDSS++V TDR+SCFSRN
Sbjct: 59 FFEEINHAGAGGIWAELVSNRGFEAGGPHTPSNINPWSIIGDDSSIFVGTDRTSCFSRNK 118
Query: 385 VALRMEVLCNDCPAGGVGIYNPGFWGMNIEDGKTYHLVMYVKSPKTTCLTVSLTSSDGLQ 564
VALRMEVLC++CP GGVGIYNPGFWGMNIEDGKTY+LVM+VKSP+T +TVSLTSSDGLQ
Sbjct: 119 VALRMEVLCDNCPVGGVGIYNPGFWGMNIEDGKTYNLVMHVKSPETIEMTVSLTSSDGLQ 178
Query: 565 NLAS 576
LAS
Sbjct: 179 VLAS 182
>sptr|Q7XRW2|Q7XRW2 OSJNBb0058J09.6 protein.
Length = 790
Score = 258 bits (660), Expect = 3e-68
Identities = 125/175 (71%), Positives = 143/175 (81%)
Frame = +1
Query: 52 RTLIHXXXXXXCIASKCLGGASGLNSTQMVTLKVDASPKLARKIPDTFLGVFFEEMGHGG 231
R +I C++ KCL S +N+TQ+ LKVDASP+ ARKIP+T G+FFEE+ H G
Sbjct: 215 RRIIFCVLLLLCVSCKCL--TSEVNTTQLAVLKVDASPQHARKIPETLFGIFFEEINHAG 272
Query: 232 AGGIWAELVSNRGFEAGGPNTPSNIDPWLIVGDDSSVYVETDRSSCFSRNIVALRMEVLC 411
AGGIWAELVSNRGFEAGGPNTPS+IDPW I+G++S + V TDRSSCFSRNI+ALRMEVLC
Sbjct: 273 AGGIWAELVSNRGFEAGGPNTPSSIDPWSIIGNESVISVATDRSSCFSRNIIALRMEVLC 332
Query: 412 NDCPAGGVGIYNPGFWGMNIEDGKTYHLVMYVKSPKTTCLTVSLTSSDGLQNLAS 576
DC AGGVGIYNPGFWGMNIEDGK Y LVMY KS + T LTVSLTSSDGLQNL+S
Sbjct: 333 GDCQAGGVGIYNPGFWGMNIEDGKNYSLVMYAKSLENTELTVSLTSSDGLQNLSS 387
>sptrnew|CAD39867|CAD39867 OSJNBb0058J09.4 protein.
Length = 654
Score = 258 bits (660), Expect = 3e-68
Identities = 125/175 (71%), Positives = 143/175 (81%)
Frame = +1
Query: 52 RTLIHXXXXXXCIASKCLGGASGLNSTQMVTLKVDASPKLARKIPDTFLGVFFEEMGHGG 231
R +I C++ KCL S +N+TQ+ LKVDASP+ ARKIP+T G+FFEE+ H G
Sbjct: 8 RRIIFCVLLLLCVSCKCL--TSEVNTTQLAVLKVDASPQHARKIPETLFGIFFEEINHAG 65
Query: 232 AGGIWAELVSNRGFEAGGPNTPSNIDPWLIVGDDSSVYVETDRSSCFSRNIVALRMEVLC 411
AGGIWAELVSNRGFEAGGPNTPS+IDPW I+G++S + V TDRSSCFSRNI+ALRMEVLC
Sbjct: 66 AGGIWAELVSNRGFEAGGPNTPSSIDPWSIIGNESVISVATDRSSCFSRNIIALRMEVLC 125
Query: 412 NDCPAGGVGIYNPGFWGMNIEDGKTYHLVMYVKSPKTTCLTVSLTSSDGLQNLAS 576
DC AGGVGIYNPGFWGMNIEDGK Y LVMY KS + T LTVSLTSSDGLQNL+S
Sbjct: 126 GDCQAGGVGIYNPGFWGMNIEDGKNYSLVMYAKSLENTELTVSLTSSDGLQNLSS 180
>sptr|Q7X9G7|Q7X9G7 Alpha-L-arabinofuranosidase.
Length = 675
Score = 225 bits (574), Expect = 3e-58
Identities = 110/169 (65%), Positives = 129/169 (76%), Gaps = 5/169 (2%)
Frame = +1
Query: 85 CIASKCLGGASGLNSTQMVTLKVDASPKLARKIPDTFLGVFFEEMGHGGAGGIWAELVSN 264
C +C A G+++ Q L +DAS AR I DT G+FFEE+ H GAGG+WAELVSN
Sbjct: 18 CSVYRCF--AIGVDANQTANLLIDASQASARPISDTLFGIFFEEINHAGAGGVWAELVSN 75
Query: 265 RGFEAGGPNTPSNIDPWLIVGDDSSVYVETDRSSCFSRNIVALRMEVLC-----NDCPAG 429
RGFEAGGPNTPSNIDPW I+G++SS+ V TDRSSCF RN VALRMEVLC N CPAG
Sbjct: 76 RGFEAGGPNTPSNIDPWAIIGNESSLIVSTDRSSCFDRNKVALRMEVLCDTQGANSCPAG 135
Query: 430 GVGIYNPGFWGMNIEDGKTYHLVMYVKSPKTTCLTVSLTSSDGLQNLAS 576
GVGIYNPGFWGMNIE GKTY+ V+YV+S + ++VSLT SDGLQ LA+
Sbjct: 136 GVGIYNPGFWGMNIEKGKTYNAVLYVRSSGSINVSVSLTGSDGLQKLAA 184
>sptrnew|BAC99303|BAC99303 Alpha-L-arabinofuranosidase.
Length = 674
Score = 223 bits (568), Expect = 1e-57
Identities = 109/169 (64%), Positives = 128/169 (75%), Gaps = 5/169 (2%)
Frame = +1
Query: 85 CIASKCLGGASGLNSTQMVTLKVDASPKLARKIPDTFLGVFFEEMGHGGAGGIWAELVSN 264
C +C A G+++ Q L +DAS AR I DT G+FFEE+ H GAGG+WAELVSN
Sbjct: 18 CSVYRCF--AIGVDANQTANLLIDASQASARPISDTLFGIFFEEINHAGAGGVWAELVSN 75
Query: 265 RGFEAGGPNTPSNIDPWLIVGDDSSVYVETDRSSCFSRNIVALRMEVLC-----NDCPAG 429
RGFEAGGPNTPSNIDPW I+G++S + V TDRSSCF RN VALRMEVLC N CPAG
Sbjct: 76 RGFEAGGPNTPSNIDPWAIIGNESFLIVSTDRSSCFDRNKVALRMEVLCDTQGANSCPAG 135
Query: 430 GVGIYNPGFWGMNIEDGKTYHLVMYVKSPKTTCLTVSLTSSDGLQNLAS 576
GVGIYNPGFWGMNIE GKTY+ V+YV+S + ++VSLT SDGLQ LA+
Sbjct: 136 GVGIYNPGFWGMNIEKGKTYNAVLYVRSSGSINVSVSLTGSDGLQKLAA 184
>sptr|Q8LIG5|Q8LIG5 Putative arabinoxylan narabinofuranohydrolase
isoenzyme AXAH-I.
Length = 664
Score = 219 bits (559), Expect = 2e-56
Identities = 105/168 (62%), Positives = 129/168 (76%), Gaps = 5/168 (2%)
Frame = +1
Query: 88 IASKCLGGASGLNSTQMVTLKVDASPKLARKIPDTFLGVFFEEMGHGGAGGIWAELVSNR 267
+ + C + G + Q L VDASP+ ARKIPD G+FFEE+ H GAGG+WAELVSNR
Sbjct: 18 VYAMCWTVSLGFVAGQTGQLNVDASPQNARKIPDKMFGIFFEEINHAGAGGLWAELVSNR 77
Query: 268 GFEAGGPNTPSNIDPWLIVGDDSSVYVETDRSSCFSRNIVALRMEVLC-----NDCPAGG 432
GFEAGGPNTPSNIDPWLI+G++SS+ V TDR+SCF +N VALRMEVLC N+CP+GG
Sbjct: 78 GFEAGGPNTPSNIDPWLIIGNESSIIVGTDRTSCFEKNPVALRMEVLCDSKGTNNCPSGG 137
Query: 433 VGIYNPGFWGMNIEDGKTYHLVMYVKSPKTTCLTVSLTSSDGLQNLAS 576
VG+YNPG+WGMNIE + Y + ++++S LTVSLTSSDGLQ LAS
Sbjct: 138 VGVYNPGYWGMNIERRRVYKVGLHIRSSDAVSLTVSLTSSDGLQKLAS 185
>sptrnew|BAC99302|BAC99302 Alpha-L-arabinofuranosidase.
Length = 674
Score = 215 bits (548), Expect = 3e-55
Identities = 100/168 (59%), Positives = 129/168 (76%), Gaps = 5/168 (2%)
Frame = +1
Query: 88 IASKCLGGASGLNSTQMVTLKVDASPKLARKIPDTFLGVFFEEMGHGGAGGIWAELVSNR 267
+++ C A+G+ + Q L V+AS AR+IPDT G+FFEE+ H GAGG+WAELV+NR
Sbjct: 18 LSALCQCSATGVEANQTAVLLVNASEASARRIPDTLFGIFFEEINHAGAGGLWAELVNNR 77
Query: 268 GFEAGGPNTPSNIDPWLIVGDDSSVYVETDRSSCFSRNIVALRMEVLC-----NDCPAGG 432
GFE GGPN PSNIDPW I+GD+S V V TDRSSCF RN +A++++VLC N CP GG
Sbjct: 78 GFEGGGPNVPSNIDPWSIIGDESKVIVSTDRSSCFDRNKIAVQVQVLCDHTGANICPDGG 137
Query: 433 VGIYNPGFWGMNIEDGKTYHLVMYVKSPKTTCLTVSLTSSDGLQNLAS 576
VGIYNPGFWGMNIE GK+Y LV+YV+S ++ ++V+LT S+GLQ LA+
Sbjct: 138 VGIYNPGFWGMNIEQGKSYKLVLYVRSEESVNVSVALTGSNGLQKLAA 185
>sptr|Q944F3|Q944F3 Arabinosidase ARA-1.
Length = 674
Score = 215 bits (548), Expect = 3e-55
Identities = 100/168 (59%), Positives = 129/168 (76%), Gaps = 5/168 (2%)
Frame = +1
Query: 88 IASKCLGGASGLNSTQMVTLKVDASPKLARKIPDTFLGVFFEEMGHGGAGGIWAELVSNR 267
+++ C A+G+ + Q L V+AS AR+IPDT G+FFEE+ H GAGG+WAELV+NR
Sbjct: 18 LSALCQCSATGVEANQTAVLLVNASEASARRIPDTLFGIFFEEINHAGAGGLWAELVNNR 77
Query: 268 GFEAGGPNTPSNIDPWLIVGDDSSVYVETDRSSCFSRNIVALRMEVLC-----NDCPAGG 432
GFE GGPN PSNIDPW I+GD+S V V TDRSSCF RN +A++++VLC N CP GG
Sbjct: 78 GFEGGGPNVPSNIDPWSIIGDESKVIVSTDRSSCFDRNKIAVQVQVLCDHTGANICPDGG 137
Query: 433 VGIYNPGFWGMNIEDGKTYHLVMYVKSPKTTCLTVSLTSSDGLQNLAS 576
VGIYNPGFWGMNIE GK+Y LV+YV+S ++ ++V+LT S+GLQ LA+
Sbjct: 138 VGIYNPGFWGMNIEQGKSYKLVLYVRSEESVNVSVALTGSNGLQKLAA 185
>sptr|Q84RC5|Q84RC5 Alpha-L-arabinofuranosidase.
Length = 678
Score = 208 bits (530), Expect = 4e-53
Identities = 100/148 (67%), Positives = 119/148 (80%), Gaps = 2/148 (1%)
Frame = +1
Query: 139 VTLKVDASPKLARKIPDTFLGVFFEEMGHGGAGGIWAELVSNRGFEAGGPNTPSNIDPWL 318
VTL+VDAS R IP+T G+FFEE+ H GAGG+WAELVSNRGFEAGG NTPSNI PW
Sbjct: 41 VTLQVDASNGGGRPIPETLFGIFFEEINHAGAGGLWAELVSNRGFEAGGQNTPSNIWPWS 100
Query: 319 IVGDDSSVYVETDRSSCFSRNIVALRMEVLCND--CPAGGVGIYNPGFWGMNIEDGKTYH 492
IVGD SS+YV TDRSSCF RN +ALRM+VLC+ CP+GGVG+YNPG+WGMNIE+GK Y
Sbjct: 101 IVGDHSSIYVATDRSSCFERNKIALRMDVLCDSKGCPSGGVGVYNPGYWGMNIEEGKKYK 160
Query: 493 LVMYVKSPKTTCLTVSLTSSDGLQNLAS 576
+ +YV+S L+VSLTSS+G + LAS
Sbjct: 161 VALYVRSTGDIDLSVSLTSSNGSRTLAS 188
>sptr|Q94JT7|Q94JT7 AT3g10740/T7M13_18.
Length = 678
Score = 208 bits (530), Expect = 4e-53
Identities = 100/148 (67%), Positives = 119/148 (80%), Gaps = 2/148 (1%)
Frame = +1
Query: 139 VTLKVDASPKLARKIPDTFLGVFFEEMGHGGAGGIWAELVSNRGFEAGGPNTPSNIDPWL 318
VTL+VDAS R IP+T G+FFEE+ H GAGG+WAELVSNRGFEAGG NTPSNI PW
Sbjct: 41 VTLQVDASNGGGRPIPETLFGIFFEEINHAGAGGLWAELVSNRGFEAGGQNTPSNIWPWS 100
Query: 319 IVGDDSSVYVETDRSSCFSRNIVALRMEVLCND--CPAGGVGIYNPGFWGMNIEDGKTYH 492
IVGD SS+YV TDRSSCF RN +ALRM+VLC+ CP+GGVG+YNPG+WGMNIE+GK Y
Sbjct: 101 IVGDHSSIYVATDRSSCFERNKIALRMDVLCDSKGCPSGGVGVYNPGYWGMNIEEGKKYK 160
Query: 493 LVMYVKSPKTTCLTVSLTSSDGLQNLAS 576
+ +YV+S L+VSLTSS+G + LAS
Sbjct: 161 VALYVRSTGDIDLSVSLTSSNGSRTLAS 188
>sptr|Q9SG80|Q9SG80 Putative alpha-L-arabinofuranosidase.
Length = 678
Score = 208 bits (530), Expect = 4e-53
Identities = 100/148 (67%), Positives = 119/148 (80%), Gaps = 2/148 (1%)
Frame = +1
Query: 139 VTLKVDASPKLARKIPDTFLGVFFEEMGHGGAGGIWAELVSNRGFEAGGPNTPSNIDPWL 318
VTL+VDAS R IP+T G+FFEE+ H GAGG+WAELVSNRGFEAGG NTPSNI PW
Sbjct: 41 VTLQVDASNGGGRPIPETLFGIFFEEINHAGAGGLWAELVSNRGFEAGGQNTPSNIWPWS 100
Query: 319 IVGDDSSVYVETDRSSCFSRNIVALRMEVLCND--CPAGGVGIYNPGFWGMNIEDGKTYH 492
IVGD SS+YV TDRSSCF RN +ALRM+VLC+ CP+GGVG+YNPG+WGMNIE+GK Y
Sbjct: 101 IVGDHSSIYVATDRSSCFERNKIALRMDVLCDSKGCPSGGVGVYNPGYWGMNIEEGKKYK 160
Query: 493 LVMYVKSPKTTCLTVSLTSSDGLQNLAS 576
+ +YV+S L+VSLTSS+G + LAS
Sbjct: 161 VALYVRSTGDIDLSVSLTSSNGSRTLAS 188
>sptr|Q8VZR2|Q8VZR2 Putative arabinosidase
(Alpha-L-arabinofuranosidase) (Hypothetical protein
At5g26120).
Length = 674
Score = 204 bits (519), Expect = 7e-52
Identities = 98/149 (65%), Positives = 117/149 (78%), Gaps = 2/149 (1%)
Frame = +1
Query: 136 MVTLKVDASPKLARKIPDTFLGVFFEEMGHGGAGGIWAELVSNRGFEAGGPNTPSNIDPW 315
+VTL+VDAS R IP+T G+FFEE+ H GAGG+WAELVSNRGFEAGG PSNI PW
Sbjct: 39 IVTLQVDASNVTRRPIPETLFGIFFEEINHAGAGGLWAELVSNRGFEAGGQIIPSNIWPW 98
Query: 316 LIVGDDSSVYVETDRSSCFSRNIVALRMEVLC--NDCPAGGVGIYNPGFWGMNIEDGKTY 489
I+GD+SS+YV TDRSSCF RN +ALRMEVLC N CP GGVG+YNPG+WGMNIE+GK Y
Sbjct: 99 SIIGDESSIYVVTDRSSCFERNKIALRMEVLCDSNSCPLGGVGVYNPGYWGMNIEEGKKY 158
Query: 490 HLVMYVKSPKTTCLTVSLTSSDGLQNLAS 576
+V+YV+S ++VS TSS+G LAS
Sbjct: 159 KVVLYVRSTGDIDVSVSFTSSNGSVTLAS 187
>sptrnew|AAR27067|AAR27067 A-arabinofuranosidase 1 (Fragment).
Length = 272
Score = 102 bits (254), Expect = 4e-21
Identities = 49/82 (59%), Positives = 61/82 (74%), Gaps = 5/82 (6%)
Frame = +1
Query: 346 VETDRSSCFSRNIVALRMEVLC-----NDCPAGGVGIYNPGFWGMNIEDGKTYHLVMYVK 510
V TDRSSCF RN +AL+MEVLC N CP GVGIYNPG+WGMNIE GK+Y +++YV+
Sbjct: 1 VSTDRSSCFERNKIALKMEVLCDSKGPNICPPEGVGIYNPGYWGMNIEQGKSYKVILYVR 60
Query: 511 SPKTTCLTVSLTSSDGLQNLAS 576
S ++V+LT S+G Q LAS
Sbjct: 61 SDDAINVSVALTGSNGSQKLAS 82
>sptr|Q9XH04|Q9XH04 T1N24.13 protein.
Length = 521
Score = 80.9 bits (198), Expect = 1e-14
Identities = 38/62 (61%), Positives = 47/62 (75%), Gaps = 2/62 (3%)
Frame = +1
Query: 397 MEVLC--NDCPAGGVGIYNPGFWGMNIEDGKTYHLVMYVKSPKTTCLTVSLTSSDGLQNL 570
MEVLC N CP GGVG+YNPG+WGMNIE+GK Y +V+YV+S ++VS TSS+G L
Sbjct: 1 MEVLCDSNSCPLGGVGVYNPGYWGMNIEEGKKYKVVLYVRSTGDIDVSVSFTSSNGSVTL 60
Query: 571 AS 576
AS
Sbjct: 61 AS 62
>sptr|Q828B3|Q828B3 Putative secreted arabinosidase.
Length = 710
Score = 60.5 bits (145), Expect = 2e-08
Identities = 52/189 (27%), Positives = 85/189 (44%), Gaps = 4/189 (2%)
Frame = +1
Query: 46 PPRTLIHXXXXXXCIASKCLGGASGLNSTQMVTLKVDASPK-LARKIPDTFLGVFFEEMG 222
P RT + +G + ++ + + PK KI DT GVFFE++
Sbjct: 3 PTRTRWRLGLTAAALLVASVGVPAPAHAEDITDYAIAVDPKGSGAKIDDTMYGVFFEDIN 62
Query: 223 HGGAGGIWAELVSNRGFE-AGGPNTP-SNIDPWLIVGDDSSVYVETDRSSCFSRNIVALR 396
GG++AELV NR FE A NT + + W G + V + R + +R+ +AL
Sbjct: 63 RAADGGLYAELVQNRSFEYATADNTSYTPLTSWNTSG-TADVVSDDGRLNARNRSYLAL- 120
Query: 397 MEVLCNDCPAGGVGIYNPGF-WGMNIEDGKTYHLVMYVKSPKTTCLTVSLTSSDGLQNLA 573
G + N G+ G+ +E GK Y ++ ++ + L+V+L +DG +LA
Sbjct: 121 ---------GGDSSVTNSGYNTGIAVESGKVYDFSVWARADQADPLSVTLHDTDG--DLA 169
Query: 574 SGYHNSCRG 600
+ RG
Sbjct: 170 RARRVTVRG 178
>sw|P82593|ABF1_STRCX Alpha-L-arabinofuranosidase I precursor (EC
3.2.1.55) (Arabinosidase I) (Alpha-L-AFase I).
Length = 825
Score = 59.3 bits (142), Expect = 4e-08
Identities = 43/142 (30%), Positives = 70/142 (49%), Gaps = 3/142 (2%)
Frame = +1
Query: 142 TLKVDASPKLARKIPDTFLGVFFEEMGHGGAGGIWAELVSNRGFEAGGPNTPS--NIDPW 315
++ VD + K A I DT GVFFE++ GG++AELV NR FE + S + W
Sbjct: 36 SITVDPAAKGAA-IDDTMYGVFFEDINRAADGGLYAELVQNRSFEYSTDDNRSYTPLTSW 94
Query: 316 LIVGDDSSVYVETDRSSCFSRNIVALRMEVLCNDCPAGGVGIYNPGF-WGMNIEDGKTYH 492
++ G V + R + +RN ++L G + N G+ G+ +E GK Y
Sbjct: 95 IVDG-TGEVVNDAGRLNERNRNYLSL----------GAGSSVTNAGYNTGIRVEQGKRYD 143
Query: 493 LVMYVKSPKTTCLTVSLTSSDG 558
++ ++ + LTV+L + G
Sbjct: 144 FSVWARAGSASTLTVALKDAAG 165
>sptr|Q8A1K4|Q8A1K4 Alpha-L-arabinofuranosidase A precursor.
Length = 825
Score = 58.9 bits (141), Expect = 5e-08
Identities = 44/156 (28%), Positives = 76/156 (48%), Gaps = 8/156 (5%)
Frame = +1
Query: 115 SGLNSTQMVTLKVDASPKLARKIPDTFLGVFFEEMGHGGAGGIWAELVSNRGFE-----A 279
+GL S V V A P+ ++I D +G+FFE++ + GG++AEL+ NR FE
Sbjct: 186 AGLKS---VKATVTAQPEETKEISDLLMGIFFEDINYSADGGLYAELIQNRDFEYEPSDR 242
Query: 280 GGPNTPSNIDPWLIVGDDSSVYVETDRSSCFSRNIVALRMEVLCNDCPAGGVGIYNPGFW 459
G ++ W + G++++ + T + A VL + P G + N GF
Sbjct: 243 EGDKNWNSTHSWKLEGENATFTISTSDPIHPNNPHYA----VLKTNQP--GAALTNTGFD 296
Query: 460 GMNIEDGKTYHLVMYVKSP---KTTCLTVSLTSSDG 558
G+ ++ G+ Y ++ + P K+ L V L +DG
Sbjct: 297 GIALKAGEKYDFSLFARIPEGSKSGKLLVRLVDADG 332
>sw|P42254|ABFA_ASPNG Alpha-L-arabinofuranosidase A precursor (EC
3.2.1.55) (Arabinosidase A) (ABF A).
Length = 628
Score = 58.5 bits (140), Expect = 6e-08
Identities = 34/118 (28%), Positives = 61/118 (51%)
Frame = +1
Query: 199 GVFFEEMGHGGAGGIWAELVSNRGFEAGGPNTPSNIDPWLIVGDDSSVYVETDRSSCFSR 378
G FE++ H G GGI+ +++ N G + P N+ W VG D+++ ++ D S S
Sbjct: 44 GFMFEDINHSGDGGIYGQMLQNPGLQGTAP----NLTAWAAVG-DATIAIDGD-SPLTSA 97
Query: 379 NIVALRMEVLCNDCPAGGVGIYNPGFWGMNIEDGKTYHLVMYVKSPKTTCLTVSLTSS 552
+++ + D G VG+ N G+WG+ + DG +H ++K + +TV L +
Sbjct: 98 IPSTIKLNIA--DDATGAVGLTNEGYWGIPV-DGSEFHSSFWIKGDYSGDITVRLVGN 152
>sptr|O88043|O88043 Putative secreted arabinosidase.
Length = 824
Score = 57.8 bits (138), Expect = 1e-07
Identities = 44/140 (31%), Positives = 65/140 (46%), Gaps = 1/140 (0%)
Frame = +1
Query: 142 TLKVDASPKLARKIPDTFLGVFFEEMGHGGAGGIWAELVSNRGFEAGGPNTPSNIDPWLI 321
T+ VD S + I DT GVF+E++ GG++AELV NR FE + S P
Sbjct: 35 TITVDPSDR-GPAIDDTMYGVFYEDINRAADGGLYAELVQNRSFEYSTADNAS-YTPLTA 92
Query: 322 VGDDSSVYVETDRSSCFSRNIVALRMEVLCNDCPAGGVGIYNPGF-WGMNIEDGKTYHLV 498
D + V D RN L + A G + N G+ G+ +E G+ Y
Sbjct: 93 WAADGTAEVVNDGGRLNERNRNYLSL--------AAGSTVTNAGYNTGIRVEKGERYDFS 144
Query: 499 MYVKSPKTTCLTVSLTSSDG 558
++ ++ T LTV+LT + G
Sbjct: 145 VWSRAGHGTTLTVTLTDAAG 164
>sptr|Q8NK90|Q8NK90 Alpha-L-arabinofuranosidase A (EC 3.2.1.55).
Length = 628
Score = 57.0 bits (136), Expect = 2e-07
Identities = 45/162 (27%), Positives = 75/162 (46%), Gaps = 7/162 (4%)
Frame = +1
Query: 88 IASKCLGGASGLN-------STQMVTLKVDASPKLARKIPDTFLGVFFEEMGHGGAGGIW 246
+A L G S L+ V+LKV S + G FE++ H G GGI+
Sbjct: 2 VAFSALSGVSALSLLLCLVQHAHGVSLKV--STQGGNSSSPILYGFMFEDINHSGDGGIY 59
Query: 247 AELVSNRGFEAGGPNTPSNIDPWLIVGDDSSVYVETDRSSCFSRNIVALRMEVLCNDCPA 426
+L+ N G + T N+ W VG D+++ ++ D S S ++++V D
Sbjct: 60 GQLLQNPGLQ----GTTPNLTAWAAVG-DATIAIDGD-SPLTSAIPSTIKLDVA--DDAT 111
Query: 427 GGVGIYNPGFWGMNIEDGKTYHLVMYVKSPKTTCLTVSLTSS 552
G VG+ N G+WG+ + DG + ++K + +TV L +
Sbjct: 112 GAVGLTNEGYWGIPV-DGSEFQSSFWIKGEYSGDITVRLVGN 152
>sptr|Q96X54|Q96X54 Alpha-L-arabinofuranosidase A (EC 3.2.1.55).
Length = 628
Score = 57.0 bits (136), Expect = 2e-07
Identities = 45/162 (27%), Positives = 75/162 (46%), Gaps = 7/162 (4%)
Frame = +1
Query: 88 IASKCLGGASGLN-------STQMVTLKVDASPKLARKIPDTFLGVFFEEMGHGGAGGIW 246
+A L G S L+ V+LKV S + G FE++ H G GGI+
Sbjct: 2 VAFSALSGVSALSLLLCLVQHAHGVSLKV--STQGGNSSSPILYGFMFEDINHSGDGGIY 59
Query: 247 AELVSNRGFEAGGPNTPSNIDPWLIVGDDSSVYVETDRSSCFSRNIVALRMEVLCNDCPA 426
+L+ N G + T N+ W VG D+++ ++ D S S ++++V D
Sbjct: 60 GQLLQNPGLQ----GTTPNLTAWAAVG-DATIAIDGD-SPLTSAIPSTIKLDVA--DDAT 111
Query: 427 GGVGIYNPGFWGMNIEDGKTYHLVMYVKSPKTTCLTVSLTSS 552
G VG+ N G+WG+ + DG + ++K + +TV L +
Sbjct: 112 GAVGLTNEGYWGIPV-DGSEFQSSFWIKGDYSGDITVRLVGN 152
>sptr|Q8AAU6|Q8AAU6 Alpha-L-arabinofuranosidase A precursor.
Length = 660
Score = 53.1 bits (126), Expect = 3e-06
Identities = 36/120 (30%), Positives = 55/120 (45%), Gaps = 3/120 (2%)
Frame = +1
Query: 166 KLARKIPDTFLGVFFEEMGHGGAGGIWAELVSNRGFEAGGPNTPSNIDPWLIVGDDSSVY 345
KL +I T G+FFE++ + GG++AELV NR FE P + W G
Sbjct: 32 KLGAEIQPTMYGLFFEDINYAADGGLYAELVKNRSFE-----FPQRLMGWKTYGK----- 81
Query: 346 VETDRSSCFSRNIVALRMEVLCNDCPAGG---VGIYNPGFWGMNIEDGKTYHLVMYVKSP 516
V F RN +R+ D P G+ N GF+G+ ++ G+ Y ++ + P
Sbjct: 82 VTLQDDGPFERNPHYVRL-----DNPGHAHKHTGLDNEGFFGIGVKQGEEYRFSVWARLP 136
>sptr|Q59218|Q59218 Arabinosidase (EC 3.2.1.55).
Length = 660
Score = 51.2 bits (121), Expect = 1e-05
Identities = 34/118 (28%), Positives = 56/118 (47%)
Frame = +1
Query: 166 KLARKIPDTFLGVFFEEMGHGGAGGIWAELVSNRGFEAGGPNTPSNIDPWLIVGDDSSVY 345
KL +I T G+FFE++ + GG++AELV NR FE P ++ W G
Sbjct: 32 KLGAEIQPTMYGLFFEDINYAADGGLYAELVKNRSFE-----FPQHLMGWKTYGK----- 81
Query: 346 VETDRSSCFSRNIVALRMEVLCNDCPAGGVGIYNPGFWGMNIEDGKTYHLVMYVKSPK 519
V F RN +R+ + G+ N GF+G+ ++ G+ Y ++ + P+
Sbjct: 82 VSLMNDGPFERNPHYVRLSDPGH--AHKHTGLDNEGFFGIGVKKGEEYRFSVWARLPQ 137
>sptr|Q8A359|Q8A359 Alpha-L-arabinofuranosidase A precursor.
Length = 709
Score = 50.8 bits (120), Expect = 1e-05
Identities = 48/175 (27%), Positives = 69/175 (39%), Gaps = 27/175 (15%)
Frame = +1
Query: 115 SGLNSTQMVTLKVDASPKLARKIPDTFLGVFFEEMGHGGAGGIWAELVSNRGFEAGGP-- 288
S +N T L +D RK+ G +EE+G G G + AELV NR FE P
Sbjct: 53 SQVNDTHEGVLHIDKQK--TRKVSRVQYGFHYEEIGMIGEGALHAELVRNRSFEEATPPA 110
Query: 289 ----------NTPS---------NIDPWLIVGDDSSVYV-----ETDRSSCFSRNIVALR 396
N P+ ++DP + Y T+ + N ++
Sbjct: 111 DLAVKNGLYQNVPNPRGKNKDVFHVDPLIGWNTYPLSYTPIFISRTEENPLNKENKYSML 170
Query: 397 MEVLCNDCPAGGVGIYNPGFWGMNIEDGKTYHLVMYVKSPK-TTCLTVSLTSSDG 558
+ V + I N G++GMN+ +YHL MY+KS T L V L G
Sbjct: 171 VNVTEDIANNPEAMILNRGYYGMNLRKEVSYHLSMYIKSKNYTALLQVMLVDEQG 225
>sptr|Q97DN6|Q97DN6 Probable alpha-arabinofuranosidase.
Length = 835
Score = 48.9 bits (115), Expect = 5e-05
Identities = 33/142 (23%), Positives = 65/142 (45%), Gaps = 3/142 (2%)
Frame = +1
Query: 142 TLKVDASPKLARKIPDTFLGVFFEEMGHGGAGGIWAELVSNRGFEAGGPNTPSNIDPWLI 321
TL ++ KL + D G+FFE++ HG GG+++EL+ N+ FE ++ W +
Sbjct: 38 TLNINTDNKLFNE-SDMLTGLFFEDINHGADGGLYSELLQNQSFE-----FKDSLSSWTV 91
Query: 322 VGDDSSVYVETDRSSCFSRNIVALRMEVLCNDCP--AGGVGIYNPGFWGMNIEDGKTYHL 495
D + + C S+ + + L +CP + + N G+ G+ + + Y
Sbjct: 92 ---DKTGSTSSTAEVCTSKPLNSNNTHYLELNCPDNNSSLKLVNSGYKGITVNNNAKYDF 148
Query: 496 VMYVKS-PKTTCLTVSLTSSDG 558
+ ++ K +T+ L +G
Sbjct: 149 YFWARNVGKGNKVTIQLEDENG 170
>sptrnew|CAF05858|CAF05858 Related to alpha-L-arabinofuranosidase A.
Length = 656
Score = 47.0 bits (110), Expect = 2e-04
Identities = 36/120 (30%), Positives = 56/120 (46%), Gaps = 2/120 (1%)
Frame = +1
Query: 199 GVFFEEMGHGGAGGIWAELVSNRGFEAGGPNT-PSNIDPWLIVGDDSSVYVETDRSSCFS 375
G+ E++ + G GGI+AEL+ NR F++G + P +D W V + + + S
Sbjct: 47 GLMHEDINNSGDGGIYAELIRNRAFQSGDESRYPVTLDGWSAV---NGAKLSLKKLSQPL 103
Query: 376 RNIVALRMEVLCNDCPAGGVGIYNPGFWGMNIEDGKTYHLVMYVKSP-KTTCLTVSLTSS 552
+ M V VG N G+WGM+++ K Y YVK + T SL S+
Sbjct: 104 SAALPYSMNVAVPSSSNKTVGFANAGYWGMDVKVQK-YTGSFYVKGAYQGKSFTASLQSA 162
>sptr|Q7S412|Q7S412 Hypothetical protein.
Length = 667
Score = 47.0 bits (110), Expect = 2e-04
Identities = 36/120 (30%), Positives = 56/120 (46%), Gaps = 2/120 (1%)
Frame = +1
Query: 199 GVFFEEMGHGGAGGIWAELVSNRGFEAGGPNT-PSNIDPWLIVGDDSSVYVETDRSSCFS 375
G+ E++ + G GGI+AEL+ NR F++G + P +D W V + + + S
Sbjct: 47 GLMHEDINNSGDGGIYAELIRNRAFQSGDESRYPVTLDGWSAV---NGAKLSLKKLSQPL 103
Query: 376 RNIVALRMEVLCNDCPAGGVGIYNPGFWGMNIEDGKTYHLVMYVKSP-KTTCLTVSLTSS 552
+ M V VG N G+WGM+++ K Y YVK + T SL S+
Sbjct: 104 SAALPYSMNVAVPSSSNKTVGFANAGYWGMDVKVQK-YTGSFYVKGAYQGKSFTASLQSA 162
>sptr|Q8G7R6|Q8G7R6 Similar to alpha-arabinofuranosidase I.
Length = 823
Score = 42.7 bits (99), Expect = 0.004
Identities = 42/157 (26%), Positives = 59/157 (37%), Gaps = 29/157 (18%)
Frame = +1
Query: 175 RKIPDTFLGVFFEEMGHGGAGGIWAELVSNRGFEAGGPNTP--SNIDPW----------- 315
RKI GVFFE++ + G GG+ +ELV N FE + P SN W
Sbjct: 17 RKISTDLWGVFFEDISYSGDGGLNSELVQNGAFEYNRADKPEWSNYTAWRKIVPAGSFAA 76
Query: 316 ---------------LIVGDDSSVYVETDRSSCFSRNIVALRMEVLCNDCPAGGVGIYNP 450
+ + V E S SR AL + C + N
Sbjct: 77 FGVGETAPVAEENPHYAIAEIGKVGGEQTADSAVSRADSALSQ--TDSACTPAAPALENL 134
Query: 451 GFWGMNIEDGKTYHLVMYVKS-PKTTCLTVSLTSSDG 558
GF GM G+TY ++ ++ K + V+L DG
Sbjct: 135 GFDGMVFRAGETYDFSIWTRAHGKALPVQVALIGDDG 171
>sptr|O15054|O15054 Hypothetical protein KIAA0346 (Fragment).
Length = 1682
Score = 37.0 bits (84), Expect = 0.20
Identities = 16/38 (42%), Positives = 23/38 (60%)
Frame = -2
Query: 299 DGVLGPPASKPLLLTSSAHIPPAPPWPISSKNTPRNVS 186
+G LGPPAS+ + T +H PP PP P +S + + S
Sbjct: 523 EGFLGPPASRFSVGTQDSHTPPTPPTPTTSSSNSNSGS 560
Database: swall
Posted date: Feb 26, 2004 3:00 PM
Number of letters in database: 439,479,560
Number of sequences in database: 1,381,838
Lambda K H
0.317 0.136 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 565,675,885
Number of Sequences: 1381838
Number of extensions: 13359443
Number of successful extensions: 44432
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 40281
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 44307
length of database: 439,479,560
effective HSP length: 117
effective length of database: 277,804,514
effective search space used: 23613383690
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)

