BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= QAF36h01.yg.2.1
(689 letters)
Database: /db/uniprot/tmp/swall
1,395,590 sequences; 442,889,342 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
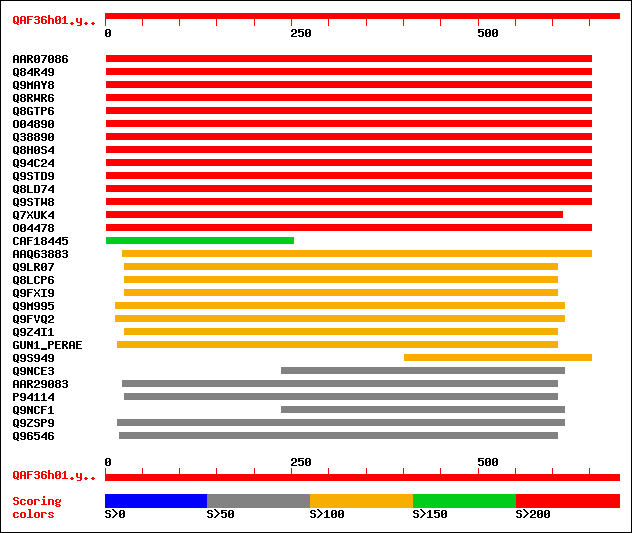
sptrnew|AAR07086|AAR07086 Putative endo-1,4-beta-glucanase. 413 e-114
sptr|Q84R49|Q84R49 Putative endo-1,4-beta-glucanase. 413 e-114
sptr|Q9MAY8|Q9MAY8 Endo-1,4-beta-glucanase Cel1. 405 e-112
sptr|Q8RWR6|Q8RWR6 Endo-1,4-beta-glucanase. 399 e-110
sptr|Q8GTP6|Q8GTP6 Endo-1,4-beta-D-glucanase (EC 3.2.1.4). 343 1e-93
sptr|O04890|O04890 Endo-1,4-beta-glucanase (EC 3.2.1.4). 335 4e-91
sptr|Q38890|Q38890 Cellulase precursor (Cellulase homolog OR16PE... 322 3e-87
sptr|Q8H0S4|Q8H0S4 At5g49720/K2I5_8. 318 3e-86
sptr|Q94C24|Q94C24 AT5g49720/K2I5_8. 318 3e-86
sptr|Q9STD9|Q9STD9 Cellulase (EC 3.2.1.4). 318 6e-86
sptr|Q8LD74|Q8LD74 Cellulase homolog OR16pep. 317 1e-85
sptr|Q9STW8|Q9STW8 Endo-1, 4-beta-glucanase like protein. 283 2e-75
sptr|Q7XUK4|Q7XUK4 OSJNBa0067K08.14 protein. 239 3e-62
sptr|O04478|O04478 F5I14.14. 210 2e-53
sptrnew|CAF18445|CAF18445 Endo-1,4-beta-D-glucanase KORRIGAN (EC... 161 7e-39
sptrnew|AAQ63883|AAQ63883 Cellulase. 137 2e-31
sptr|Q9LR07|Q9LR07 F10A5.13 (Hypothetical protein) (Putative end... 126 3e-28
sptr|Q8LCP6|Q8LCP6 Endo-beta-1,4-glucanase, putative. 125 4e-28
sptr|Q9FXI9|Q9FXI9 F6F9.1 protein (At1g19940/F6F9_1). 125 7e-28
sptr|Q9M995|Q9M995 F27J15.28 (Putative glycosyl hydrolase family... 110 2e-23
sptr|Q9FVQ2|Q9FVQ2 Endo-beta-1,4-glucanase, putative. 110 2e-23
sptr|Q9Z4I1|Q9Z4I1 Cellulase precursor (EC 3.2.1.4). 107 2e-22
sw|P05522|GUN1_PERAE Endoglucanase 1 precursor (EC 3.2.1.4) (End... 102 4e-21
sptr|Q9S949|Q9S949 CEL3=CELLULASE 3 (Fragment). 101 8e-21
sptr|Q9NCE3|Q9NCE3 Beta-1,4-endoglucanase 1. 99 4e-20
sptrnew|AAR29083|AAR29083 Cellulase (EC 3.2.1.4). 99 5e-20
sptr|P94114|P94114 Endo-beta-1,4-glucanase (EC 3.2.1.4) (Cellula... 99 5e-20
sptr|Q9NCF1|Q9NCF1 Beta-1,4-glucanase 3 (Fragment). 99 7e-20
sptr|Q9ZSP9|Q9ZSP9 Endo-beta-1,4-D-glucanase. 97 2e-19
sptr|Q96546|Q96546 Endo-beta-1,4-glucanase (EC 3.2.1.4). 97 2e-19
>sptrnew|AAR07086|AAR07086 Putative endo-1,4-beta-glucanase.
Length = 620
Score = 413 bits (1061), Expect = e-114
Identities = 193/217 (88%), Positives = 200/217 (92%)
Frame = -2
Query: 652 EXIDYXXXXXECHSCSDLAXXXXAXXXXASIVFKDSKTYSDKLVKGAKALYKFGRLQRGR 473
E IDY ECHSCSDLA A ASIVFKDSKTYSDKLV+GAKALYKFGRLQRGR
Sbjct: 251 EDIDYPRPVTECHSCSDLASEMAAALAAASIVFKDSKTYSDKLVRGAKALYKFGRLQRGR 310
Query: 472 YSPNGSDQAIFYNSTSYWDEFVWGGAWMYFATGNNSYLTVATAPGMAKHAGAYWFGSPNY 293
YSPNGSDQAIFYNSTSYWDEFVWGGAWMYFATGNN+YL+VATAPGMAKHAGAYW SPNY
Sbjct: 311 YSPNGSDQAIFYNSTSYWDEFVWGGAWMYFATGNNTYLSVATAPGMAKHAGAYWLDSPNY 370
Query: 292 GVFTWDDKLPGAQVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPVFNSFNFTKGG 113
GVFTWDDKLPGAQVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLP++NSFNFTKGG
Sbjct: 371 GVFTWDDKLPGAQVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPMYNSFNFTKGG 430
Query: 112 LIQLNHGRPQPLQYAVNAAFLASLYSDYLEAADTPGW 2
+IQLNHGRPQPLQY VNAAFLASLYSDYL+AADTPGW
Sbjct: 431 MIQLNHGRPQPLQYVVNAAFLASLYSDYLDAADTPGW 467
>sptr|Q84R49|Q84R49 Putative endo-1,4-beta-glucanase.
Length = 620
Score = 413 bits (1061), Expect = e-114
Identities = 193/217 (88%), Positives = 200/217 (92%)
Frame = -2
Query: 652 EXIDYXXXXXECHSCSDLAXXXXAXXXXASIVFKDSKTYSDKLVKGAKALYKFGRLQRGR 473
E IDY ECHSCSDLA A ASIVFKDSKTYSDKLV+GAKALYKFGRLQRGR
Sbjct: 251 EDIDYPRPVTECHSCSDLASEMAAALAAASIVFKDSKTYSDKLVRGAKALYKFGRLQRGR 310
Query: 472 YSPNGSDQAIFYNSTSYWDEFVWGGAWMYFATGNNSYLTVATAPGMAKHAGAYWFGSPNY 293
YSPNGSDQAIFYNSTSYWDEFVWGGAWMYFATGNN+YL+VATAPGMAKHAGAYW SPNY
Sbjct: 311 YSPNGSDQAIFYNSTSYWDEFVWGGAWMYFATGNNTYLSVATAPGMAKHAGAYWLDSPNY 370
Query: 292 GVFTWDDKLPGAQVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPVFNSFNFTKGG 113
GVFTWDDKLPGAQVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLP++NSFNFTKGG
Sbjct: 371 GVFTWDDKLPGAQVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPMYNSFNFTKGG 430
Query: 112 LIQLNHGRPQPLQYAVNAAFLASLYSDYLEAADTPGW 2
+IQLNHGRPQPLQY VNAAFLASLYSDYL+AADTPGW
Sbjct: 431 MIQLNHGRPQPLQYVVNAAFLASLYSDYLDAADTPGW 467
>sptr|Q9MAY8|Q9MAY8 Endo-1,4-beta-glucanase Cel1.
Length = 621
Score = 405 bits (1042), Expect = e-112
Identities = 189/217 (87%), Positives = 196/217 (90%)
Frame = -2
Query: 652 EXIDYXXXXXECHSCSDLAXXXXAXXXXASIVFKDSKTYSDKLVKGAKALYKFGRLQRGR 473
E IDY ECHSCSDLA A ASIVFKDSK YSDKLV GAKALYKFGRLQRGR
Sbjct: 251 EDIDYKRPVIECHSCSDLAAEMAAALAAASIVFKDSKAYSDKLVHGAKALYKFGRLQRGR 310
Query: 472 YSPNGSDQAIFYNSTSYWDEFVWGGAWMYFATGNNSYLTVATAPGMAKHAGAYWFGSPNY 293
YSPNGSDQ++FYNSTSYWDEFVWGGAWMYFATGN SYLT+ATAPGMAKHAGA+W GSPNY
Sbjct: 311 YSPNGSDQSLFYNSTSYWDEFVWGGAWMYFATGNTSYLTIATAPGMAKHAGAFWIGSPNY 370
Query: 292 GVFTWDDKLPGAQVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPVFNSFNFTKGG 113
GVFTWDDKLPG+QVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPVFNSFNFTKGG
Sbjct: 371 GVFTWDDKLPGSQVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPVFNSFNFTKGG 430
Query: 112 LIQLNHGRPQPLQYAVNAAFLASLYSDYLEAADTPGW 2
LIQLNHG PQPLQY VNAAFLASLY+DYL+ ADTPGW
Sbjct: 431 LIQLNHGGPQPLQYVVNAAFLASLYADYLDTADTPGW 467
>sptr|Q8RWR6|Q8RWR6 Endo-1,4-beta-glucanase.
Length = 620
Score = 399 bits (1025), Expect = e-110
Identities = 187/217 (86%), Positives = 194/217 (89%)
Frame = -2
Query: 652 EXIDYXXXXXECHSCSDLAXXXXAXXXXASIVFKDSKTYSDKLVKGAKALYKFGRLQRGR 473
E IDY ECHSCSDLA A ASIVFKDSK YSDKLV GAKALYKFGRLQRGR
Sbjct: 251 EDIDYKRPVIECHSCSDLAAGMAAALAAASIVFKDSKAYSDKLVHGAKALYKFGRLQRGR 310
Query: 472 YSPNGSDQAIFYNSTSYWDEFVWGGAWMYFATGNNSYLTVATAPGMAKHAGAYWFGSPNY 293
YSPNGSDQ++F NSTSYWDEFVWGGAWMYFATGN SYLT+ATAPGMAKHAGA+W GSPNY
Sbjct: 311 YSPNGSDQSLFCNSTSYWDEFVWGGAWMYFATGNTSYLTIATAPGMAKHAGAFWIGSPNY 370
Query: 292 GVFTWDDKLPGAQVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPVFNSFNFTKGG 113
GVFTWDDKLPG+QVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPVFNSFNFTKGG
Sbjct: 371 GVFTWDDKLPGSQVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPVFNSFNFTKGG 430
Query: 112 LIQLNHGRPQPLQYAVNAAFLASLYSDYLEAADTPGW 2
LIQLNHG PQPLQY VNAAFLASLY+DYL+ ADTP W
Sbjct: 431 LIQLNHGGPQPLQYVVNAAFLASLYADYLDTADTPRW 467
>sptr|Q8GTP6|Q8GTP6 Endo-1,4-beta-D-glucanase (EC 3.2.1.4).
Length = 621
Score = 343 bits (880), Expect = 1e-93
Identities = 162/218 (74%), Positives = 181/218 (83%), Gaps = 1/218 (0%)
Frame = -2
Query: 652 EXIDYXXXXXECHSCSDLAXXXXAXXXXASIVFKDSKTYSDKLVKGAKALYKFGRLQRGR 473
E IDY +CHSCSDLA A +SIVFKD+K YS KLV GAK L++F R QRGR
Sbjct: 253 EDIDYQRPVSQCHSCSDLAAEMAAALAASSIVFKDNKAYSQKLVHGAKTLFRFSREQRGR 312
Query: 472 YSPNGS-DQAIFYNSTSYWDEFVWGGAWMYFATGNNSYLTVATAPGMAKHAGAYWFGSPN 296
YS GS D A FYNSTSYWDEF+WGGAWMY+ATGN+SYL +AT PG+AKHAGA+W G P+
Sbjct: 313 YSAGGSTDAATFYNSTSYWDEFIWGGAWMYYATGNSSYLQLATTPGLAKHAGAFW-GGPD 371
Query: 295 YGVFTWDDKLPGAQVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPVFNSFNFTKG 116
YGV +WD+KL GAQVLLSRL LFLSPGYPYEEILRTFHNQT +MCSYLPVF +FN TKG
Sbjct: 372 YGVLSWDNKLAGAQVLLSRLXLFLSPGYPYEEILRTFHNQTSIIMCSYLPVFTTFNRTKG 431
Query: 115 GLIQLNHGRPQPLQYAVNAAFLASLYSDYLEAADTPGW 2
GLIQLNHGRPQPLQY VNAAFLA+LYS+YL+AADTPGW
Sbjct: 432 GLIQLNHGRPQPLQYVVNAAFLATLYSEYLDAADTPGW 469
>sptr|O04890|O04890 Endo-1,4-beta-glucanase (EC 3.2.1.4).
Length = 617
Score = 335 bits (858), Expect = 4e-91
Identities = 157/217 (72%), Positives = 177/217 (81%)
Frame = -2
Query: 652 EXIDYXXXXXECHSCSDLAXXXXAXXXXASIVFKDSKTYSDKLVKGAKALYKFGRLQRGR 473
E IDY ECH CSDLA A ASIVFKD+K YS KLV GA+ L+KF R QRGR
Sbjct: 251 EDIDYARPVTECHGCSDLAAEMAAALASASIVFKDNKAYSQKLVHGARTLFKFSRDQRGR 310
Query: 472 YSPNGSDQAIFYNSTSYWDEFVWGGAWMYFATGNNSYLTVATAPGMAKHAGAYWFGSPNY 293
YS G++ FYNST YWDEF+WG AW+Y+ATGN+SYL +AT PG+AKHAGA+W G P+Y
Sbjct: 311 YSV-GNEAETFYNSTGYWDEFIWGAAWLYYATGNSSYLQLATTPGIAKHAGAFW-GGPDY 368
Query: 292 GVFTWDDKLPGAQVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPVFNSFNFTKGG 113
GV +WD+KL GAQVLLSR+RLFLSPGYPYEEILRTFHNQT +MCSYLP+F SFN TKGG
Sbjct: 369 GVLSWDNKLTGAQVLLSRMRLFLSPGYPYEEILRTFHNQTSIIMCSYLPIFTSFNRTKGG 428
Query: 112 LIQLNHGRPQPLQYAVNAAFLASLYSDYLEAADTPGW 2
LIQLNHGRPQPLQY VNAAFLA+L+SDYL AADTPGW
Sbjct: 429 LIQLNHGRPQPLQYVVNAAFLATLFSDYLAAADTPGW 465
>sptr|Q38890|Q38890 Cellulase precursor (Cellulase homolog OR16PEP
precursor).
Length = 621
Score = 322 bits (825), Expect = 3e-87
Identities = 152/218 (69%), Positives = 175/218 (80%), Gaps = 1/218 (0%)
Frame = -2
Query: 652 EXIDYXXXXXECHS-CSDLAXXXXAXXXXASIVFKDSKTYSDKLVKGAKALYKFGRLQRG 476
E +DY C+ CSDLA A ASIVFKD+K YS KLV GAK +Y+FGR +RG
Sbjct: 251 EDMDYKRPVTTCNGGCSDLAAEMAAALASASIVFKDNKEYSKKLVHGAKVVYQFGRTRRG 310
Query: 475 RYSPNGSDQAIFYNSTSYWDEFVWGGAWMYFATGNNSYLTVATAPGMAKHAGAYWFGSPN 296
RYS ++ + FYNS+ YWDEF+WGGAWMY+ATGN +YL + T P MAKHAGA+W G P
Sbjct: 311 RYSAGTAESSKFYNSSMYWDEFIWGGAWMYYATGNVTYLNLITQPTMAKHAGAFW-GGPY 369
Query: 295 YGVFTWDDKLPGAQVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPVFNSFNFTKG 116
YGVF+WD+KL GAQ+LLSRLRLFLSPGYPYEEILRTFHNQT VMCSYLP+FN FN T G
Sbjct: 370 YGVFSWDNKLAGAQLLLSRLRLFLSPGYPYEEILRTFHNQTSIVMCSYLPIFNKFNRTNG 429
Query: 115 GLIQLNHGRPQPLQYAVNAAFLASLYSDYLEAADTPGW 2
GLI+LNHG PQPLQY+VNAAFLA+LYSDYL+AADTPGW
Sbjct: 430 GLIELNHGAPQPLQYSVNAAFLATLYSDYLDAADTPGW 467
>sptr|Q8H0S4|Q8H0S4 At5g49720/K2I5_8.
Length = 621
Score = 318 bits (816), Expect = 3e-86
Identities = 151/218 (69%), Positives = 174/218 (79%), Gaps = 1/218 (0%)
Frame = -2
Query: 652 EXIDYXXXXXECHS-CSDLAXXXXAXXXXASIVFKDSKTYSDKLVKGAKALYKFGRLQRG 476
E +DY C+ CSDLA A ASIVFKD+K YS KLV GAK +Y+FGR +RG
Sbjct: 251 EDMDYKRPVTTCNGGCSDLAAEMAAALASASIVFKDNKEYSKKLVHGAKVVYQFGRTRRG 310
Query: 475 RYSPNGSDQAIFYNSTSYWDEFVWGGAWMYFATGNNSYLTVATAPGMAKHAGAYWFGSPN 296
RYS ++ + FYNS+ YWDEF+WGGAWMY+ATGN +YL + T P MAKHAGA+W G P
Sbjct: 311 RYSAGTAESSKFYNSSMYWDEFIWGGAWMYYATGNVTYLNLITQPTMAKHAGAFW-GGPY 369
Query: 295 YGVFTWDDKLPGAQVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPVFNSFNFTKG 116
YGVF+WD+KL GAQ+LLSRLRLFLSPG PYEEILRTFHNQT VMCSYLP+FN FN T G
Sbjct: 370 YGVFSWDNKLAGAQLLLSRLRLFLSPGCPYEEILRTFHNQTSIVMCSYLPIFNKFNRTNG 429
Query: 115 GLIQLNHGRPQPLQYAVNAAFLASLYSDYLEAADTPGW 2
GLI+LNHG PQPLQY+VNAAFLA+LYSDYL+AADTPGW
Sbjct: 430 GLIELNHGAPQPLQYSVNAAFLATLYSDYLDAADTPGW 467
>sptr|Q94C24|Q94C24 AT5g49720/K2I5_8.
Length = 621
Score = 318 bits (816), Expect = 3e-86
Identities = 151/218 (69%), Positives = 174/218 (79%), Gaps = 1/218 (0%)
Frame = -2
Query: 652 EXIDYXXXXXECHS-CSDLAXXXXAXXXXASIVFKDSKTYSDKLVKGAKALYKFGRLQRG 476
E +DY C+ CSDLA A ASIVFKD+K YS KLV GAK +Y+FGR +RG
Sbjct: 251 EDMDYKRPVTTCNGGCSDLAAEMAAALASASIVFKDNKEYSKKLVHGAKVVYQFGRTRRG 310
Query: 475 RYSPNGSDQAIFYNSTSYWDEFVWGGAWMYFATGNNSYLTVATAPGMAKHAGAYWFGSPN 296
RYS ++ + FYNS+ YWDEF+WGGAWMY+ATGN +YL + T P MAKHAGA+W G P
Sbjct: 311 RYSAGTAESSKFYNSSMYWDEFIWGGAWMYYATGNVTYLNLITQPTMAKHAGAFW-GGPY 369
Query: 295 YGVFTWDDKLPGAQVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPVFNSFNFTKG 116
YGVF+WD+KL GAQ+LLSRLRLFLSPG PYEEILRTFHNQT VMCSYLP+FN FN T G
Sbjct: 370 YGVFSWDNKLAGAQLLLSRLRLFLSPGCPYEEILRTFHNQTSIVMCSYLPIFNKFNRTNG 429
Query: 115 GLIQLNHGRPQPLQYAVNAAFLASLYSDYLEAADTPGW 2
GLI+LNHG PQPLQY+VNAAFLA+LYSDYL+AADTPGW
Sbjct: 430 GLIELNHGAPQPLQYSVNAAFLATLYSDYLDAADTPGW 467
>sptr|Q9STD9|Q9STD9 Cellulase (EC 3.2.1.4).
Length = 621
Score = 318 bits (814), Expect = 6e-86
Identities = 150/218 (68%), Positives = 174/218 (79%), Gaps = 1/218 (0%)
Frame = -2
Query: 652 EXIDYXXXXXECHS-CSDLAXXXXAXXXXASIVFKDSKTYSDKLVKGAKALYKFGRLQRG 476
E +DY C+ CSDLA A ASIVFKD++ YS KLV GAK +Y+FGR +RG
Sbjct: 251 EDMDYKRPVTTCNGGCSDLAAEMAAALASASIVFKDNREYSKKLVHGAKTVYQFGRTRRG 310
Query: 475 RYSPNGSDQAIFYNSTSYWDEFVWGGAWMYFATGNNSYLTVATAPGMAKHAGAYWFGSPN 296
RYS ++ A FYNS+ YWDEF+WGGAW+Y+ATGN +YL + T P MAKHAGA+W G P
Sbjct: 311 RYSAGTAESAKFYNSSMYWDEFIWGGAWLYYATGNVTYLDLITKPTMAKHAGAFW-GGPY 369
Query: 295 YGVFTWDDKLPGAQVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPVFNSFNFTKG 116
YGVF+WD+KL GAQ+LLSRLRLFLSPGYPYEEI+RTFHNQT VMCSYLP FN FN T+G
Sbjct: 370 YGVFSWDNKLAGAQLLLSRLRLFLSPGYPYEEIVRTFHNQTSIVMCSYLPYFNKFNRTRG 429
Query: 115 GLIQLNHGRPQPLQYAVNAAFLASLYSDYLEAADTPGW 2
GLI+LNHG PQPLQYA NAAFLA+LYSDYL+AADTPGW
Sbjct: 430 GLIELNHGDPQPLQYAANAAFLATLYSDYLDAADTPGW 467
>sptr|Q8LD74|Q8LD74 Cellulase homolog OR16pep.
Length = 621
Score = 317 bits (811), Expect = 1e-85
Identities = 151/218 (69%), Positives = 174/218 (79%), Gaps = 1/218 (0%)
Frame = -2
Query: 652 EXIDYXXXXXECHS-CSDLAXXXXAXXXXASIVFKDSKTYSDKLVKGAKALYKFGRLQRG 476
E +DY C+ CSDLA A ASIVFKD+K YS KLV GAK +Y+FGR +RG
Sbjct: 251 EDMDYKRPVTTCNGGCSDLAAEMAAALASASIVFKDNKEYSKKLVHGAKVVYQFGRTRRG 310
Query: 475 RYSPNGSDQAIFYNSTSYWDEFVWGGAWMYFATGNNSYLTVATAPGMAKHAGAYWFGSPN 296
RYS ++ + FYNS+ YWDEF+WGGA MY+ATGN +YL + T P MAKHAGA+W G P
Sbjct: 311 RYSAGTAESSKFYNSSMYWDEFIWGGARMYYATGNVTYLNLITQPTMAKHAGAFW-GGPY 369
Query: 295 YGVFTWDDKLPGAQVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPVFNSFNFTKG 116
YGVF+WD+KL GAQ+LLSRLRLFLSPGYPYEEILRTFHNQT VMCSYLP+FN FN T G
Sbjct: 370 YGVFSWDNKLAGAQLLLSRLRLFLSPGYPYEEILRTFHNQTSIVMCSYLPIFNKFNRTNG 429
Query: 115 GLIQLNHGRPQPLQYAVNAAFLASLYSDYLEAADTPGW 2
GLI+LNHG PQPLQY+VNAAFLA+LYSDYL+AADTPGW
Sbjct: 430 GLIELNHGAPQPLQYSVNAAFLATLYSDYLDAADTPGW 467
>sptr|Q9STW8|Q9STW8 Endo-1, 4-beta-glucanase like protein.
Length = 620
Score = 283 bits (724), Expect = 2e-75
Identities = 136/218 (62%), Positives = 162/218 (74%), Gaps = 1/218 (0%)
Frame = -2
Query: 652 EXIDYXXXXXECHS-CSDLAXXXXAXXXXASIVFKDSKTYSDKLVKGAKALYKFGRLQRG 476
E I Y +C+S CSDLA A ASIVFKD++ YS LV GAK LY+F R
Sbjct: 252 EDIHYKRTVSQCYSSCSDLAAEMAAALASASIVFKDNRLYSKNLVHGAKTLYRFATTSRN 311
Query: 475 RYSPNGSDQAIFYNSTSYWDEFVWGGAWMYFATGNNSYLTVATAPGMAKHAGAYWFGSPN 296
RYS NG + + FYNS+ + DE +WGGAW+Y+ATGN +YL T+ MA+ AGA+ SP
Sbjct: 312 RYSQNGKESSKFYNSSMFEDELLWGGAWLYYATGNVTYLERVTSHHMAEKAGAFG-NSPY 370
Query: 295 YGVFTWDDKLPGAQVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPVFNSFNFTKG 116
YGVF+WD+KLPGAQ+LL+R+RLFLSPGYPYE++L FHNQT VMCSYLP + FN T G
Sbjct: 371 YGVFSWDNKLPGAQLLLTRMRLFLSPGYPYEDMLSEFHNQTGRVMCSYLPYYKKFNRTNG 430
Query: 115 GLIQLNHGRPQPLQYAVNAAFLASLYSDYLEAADTPGW 2
GLIQLNHG PQPLQY NAAFLA+L+SDYLEAADTPGW
Sbjct: 431 GLIQLNHGAPQPLQYVANAAFLAALFSDYLEAADTPGW 468
>sptr|Q7XUK4|Q7XUK4 OSJNBa0067K08.14 protein.
Length = 623
Score = 239 bits (610), Expect = 3e-62
Identities = 114/206 (55%), Positives = 146/206 (70%), Gaps = 2/206 (0%)
Frame = -2
Query: 613 SCSDLAXXXXAXXXXASIVFKDSKTYSDKLVKGAKALYKFGRL--QRGRYSPNGSDQAIF 440
S DL A ASIVF+D+ YS KLV GA A+YKF R +R YS +
Sbjct: 263 SAPDLGGEMAAALAAASIVFRDNAAYSKKLVNGAAAVYKFARSSGRRTPYSRGNQYIEYY 322
Query: 439 YNSTSYWDEFVWGGAWMYFATGNNSYLTVATAPGMAKHAGAYWFGSPNYGVFTWDDKLPG 260
YNSTSYWDE++W AWMY+ATGNN+Y+T AT P + K+A A+ + ++ VF+WD+KLPG
Sbjct: 323 YNSTSYWDEYMWSAAWMYYATGNNTYITFATDPRLPKNAKAF-YSILDFSVFSWDNKLPG 381
Query: 259 AQVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPVFNSFNFTKGGLIQLNHGRPQP 80
A++LLSRLR+FL+PGYPYEE L +HN T MC+Y P F +FNFTKGGL Q NHG+ QP
Sbjct: 382 AELLLSRLRMFLNPGYPYEESLIGYHNTTSMNMCTYFPRFGAFNFTKGGLAQFNHGKGQP 441
Query: 79 LQYAVNAAFLASLYSDYLEAADTPGW 2
LQY V +FLA+LY+DY+E+ + PGW
Sbjct: 442 LQYTVANSFLAALYADYMESVNVPGW 467
>sptr|O04478|O04478 F5I14.14.
Length = 623
Score = 210 bits (534), Expect = 2e-53
Identities = 104/219 (47%), Positives = 139/219 (63%), Gaps = 2/219 (0%)
Frame = -2
Query: 652 EXIDYXXXXXECHSCSDLAXXXXAXXXXASIVFKDSKTYSDKLVKGAKALYKFGRLQ--R 479
E + Y S +DL A ASIVF D Y+ KL KGA+ LY F R + R
Sbjct: 254 EDMSYDRPVLSSTSAADLGAEVSAALAAASIVFTDKPDYAKKLKKGAETLYPFFRSKSRR 313
Query: 478 GRYSPNGSDQAIFYNSTSYWDEFVWGGAWMYFATGNNSYLTVATAPGMAKHAGAYWFGSP 299
RYS FYNSTS +DEF+W GAW+Y+ATGN +Y+ AT P + + A A+ P
Sbjct: 314 KRYSDGQPTAQAFYNSTSMFDEFMWAGAWLYYATGNKTYIQFATTPSVPQTAKAF-ANRP 372
Query: 298 NYGVFTWDDKLPGAQVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPVFNSFNFTK 119
V +W++KLPGA +L++R RLFL+PG+PYE +L +HN T MC+YL +N FN T
Sbjct: 373 ELMVPSWNNKLPGAMLLMTRYRLFLNPGFPYENMLNRYHNATGITMCAYLKQYNVFNRTS 432
Query: 118 GGLIQLNHGRPQPLQYAVNAAFLASLYSDYLEAADTPGW 2
GGL+QLN G+P+PL+Y +A+FLASL++DYL + PGW
Sbjct: 433 GGLMQLNLGKPRPLEYVAHASFLASLFADYLNSTGVPGW 471
>sptrnew|CAF18445|CAF18445 Endo-1,4-beta-D-glucanase KORRIGAN (EC
3.2.1.4) (Fragment).
Length = 229
Score = 161 bits (408), Expect = 7e-39
Identities = 76/84 (90%), Positives = 79/84 (94%)
Frame = -2
Query: 253 VLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPVFNSFNFTKGGLIQLNHGRPQPLQ 74
VLLSRLRLFLSPGYPYEEILRTFHNQT +MCSYLPVF SFN TKGGLIQLNHGRPQPLQ
Sbjct: 1 VLLSRLRLFLSPGYPYEEILRTFHNQTSIIMCSYLPVFTSFNRTKGGLIQLNHGRPQPLQ 60
Query: 73 YAVNAAFLASLYSDYLEAADTPGW 2
Y VNAAFLA+LYSDYL+AADTPGW
Sbjct: 61 YVVNAAFLATLYSDYLDAADTPGW 84
>sptrnew|AAQ63883|AAQ63883 Cellulase.
Length = 601
Score = 137 bits (344), Expect = 2e-31
Identities = 85/219 (38%), Positives = 120/219 (54%), Gaps = 9/219 (4%)
Frame = -2
Query: 652 EXIDYXXXXXECH-SCSDLAXXXXAXXXXASIVFKDSKTYSDKLVKGAKALY----KFGR 488
E + Y C S +DLA A AS+VF++ K YS KLV+ A++LY K
Sbjct: 244 EDMSYGRPVSVCDGSATDLAGEIVAALSAASMVFEEDKEYSGKLVQAAESLYEVVTKEDP 303
Query: 487 LQRGRYSPN---GSDQAIFYNSTSYWDEFVWGGAWMYFATGNNSYLTVATAPGMAKHAGA 317
++G Y+ G + YNSTSY DE WG W++ AT N YL AT ++
Sbjct: 304 KKQGTYTAVDACGKQARMLYNSTSYKDELAWGATWLFLATKNTDYLANATQFFLSAKKDE 363
Query: 316 YWFGSPNYGVFTWDDKLPGAQVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPVFN 137
+ + GVF W++KL VLL+ +R F PG+PYE++L+ N T ++MCSYL F
Sbjct: 364 T---NLDKGVFYWNNKLSAVAVLLTGIRYFRDPGFPYEDVLKFSSNSTHSLMCSYL--FK 418
Query: 136 SF-NFTKGGLIQLNHGRPQPLQYAVNAAFLASLYSDYLE 23
+ + T GGL+ LQYAV A+FL+ LYSDY++
Sbjct: 419 KYMSRTPGGLVIPKPDNGPLLQYAVTASFLSKLYSDYID 457
>sptr|Q9LR07|Q9LR07 F10A5.13 (Hypothetical protein) (Putative
endo-beta-1,4-glucanase).
Length = 525
Score = 126 bits (316), Expect = 3e-28
Identities = 80/197 (40%), Positives = 112/197 (56%), Gaps = 3/197 (1%)
Frame = -2
Query: 607 SDLAXXXXAXXXXASIVFKDSK-TYSDKLVKGAKALYKFGRLQRGRYSPNGSDQAIFYNS 431
+++A A AS+VFKDS TYS L+K AK L+ F +RG YS N + FYNS
Sbjct: 201 TEVAAETAAAMASASLVFKDSDPTYSATLLKHAKQLFNFADTKRGSYSVNIPEVQKFYNS 260
Query: 430 TSYWDEFVWGGAWMYFATGNNSYLTVATAPGMAKHAGAYWFGSPNYGVFTWDDKLPGAQV 251
T Y DE +W +W+Y AT + +YL + G + FG+P + F+WD+KL G QV
Sbjct: 261 TGYGDELLWAASWLYHATEDKTYLDYVSNHGKEFAS----FGNPTW--FSWDNKLAGTQV 314
Query: 250 LLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLP--VFNSFNFTKGGLIQLNHGRPQPL 77
LLSRL LF + L + N VMC LP ++ + T GGLI ++ +
Sbjct: 315 LLSRL-LFFKKDLSGSKGLGNYRNTAKAVMCGLLPKSPTSTASRTNGGLIWVSEW--NSM 371
Query: 76 QYAVNAAFLASLYSDYL 26
Q +V++AFLASL+SDY+
Sbjct: 372 QQSVSSAFLASLFSDYM 388
>sptr|Q8LCP6|Q8LCP6 Endo-beta-1,4-glucanase, putative.
Length = 525
Score = 125 bits (315), Expect = 4e-28
Identities = 80/197 (40%), Positives = 112/197 (56%), Gaps = 3/197 (1%)
Frame = -2
Query: 607 SDLAXXXXAXXXXASIVFKDSK-TYSDKLVKGAKALYKFGRLQRGRYSPNGSDQAIFYNS 431
+++A A AS+VFKDS TYS L+K AK L+ F +RG YS N + FYNS
Sbjct: 201 TEVAAETAAAMASASLVFKDSDPTYSATLLKHAKQLFDFADTKRGSYSVNIPEVQKFYNS 260
Query: 430 TSYWDEFVWGGAWMYFATGNNSYLTVATAPGMAKHAGAYWFGSPNYGVFTWDDKLPGAQV 251
T Y DE +W +W+Y AT + +YL + G + FG+P + F+WD+KL G QV
Sbjct: 261 TGYGDELLWAASWLYHATEDKTYLDYVSNHGKEFAS----FGNPTW--FSWDNKLAGTQV 314
Query: 250 LLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLP--VFNSFNFTKGGLIQLNHGRPQPL 77
LLSRL LF + L + N VMC LP ++ + T GGLI ++ +
Sbjct: 315 LLSRL-LFFKKDLSGSKGLGNYRNTAKAVMCGLLPKSPTSTASRTNGGLIWVSEW--NSM 371
Query: 76 QYAVNAAFLASLYSDYL 26
Q +V++AFLASL+SDY+
Sbjct: 372 QQSVSSAFLASLFSDYM 388
>sptr|Q9FXI9|Q9FXI9 F6F9.1 protein (At1g19940/F6F9_1).
Length = 515
Score = 125 bits (313), Expect = 7e-28
Identities = 79/197 (40%), Positives = 113/197 (57%), Gaps = 3/197 (1%)
Frame = -2
Query: 607 SDLAXXXXAXXXXASIVFKDSKT-YSDKLVKGAKALYKFGRLQRGRYSPNGSDQAIFYNS 431
+++A A AS+VFK+S T YS L+K AK L+ F RG YS N + +YNS
Sbjct: 192 TEVAAETAAAMAAASLVFKESDTKYSSTLLKHAKQLFDFADNNRGSYSVNIPEVQSYYNS 251
Query: 430 TSYWDEFVWGGAWMYFATGNNSYLTVATAPGMAKHAGAYWFGSPNYGVFTWDDKLPGAQV 251
T Y DE +W +W+Y AT + +YL + G + G FGSP++ F+WD+KLPG +
Sbjct: 252 TGYGDELLWAASWLYHATEDQTYLDFVSENG--EEFGN--FGSPSW--FSWDNKLPGTHI 305
Query: 250 LLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLP--VFNSFNFTKGGLIQLNHGRPQPL 77
LLSRL F G + L+ F + VMC +P + + T GGLI ++ L
Sbjct: 306 LLSRL-TFFKKGLSGSKGLQGFKETAEAVMCGLIPSSPTATSSRTDGGLIWVSEW--NAL 362
Query: 76 QYAVNAAFLASLYSDYL 26
Q+ V++AFLA+LYSDY+
Sbjct: 363 QHPVSSAFLATLYSDYM 379
>sptr|Q9M995|Q9M995 F27J15.28 (Putative glycosyl hydrolase family 9
(Endo-1,4-beta-glucanase) protein).
Length = 627
Score = 110 bits (274), Expect = 2e-23
Identities = 73/206 (35%), Positives = 105/206 (50%), Gaps = 5/206 (2%)
Frame = -2
Query: 616 HSCSDLAXXXXAXXXXASIVFKD-SKTYSDKLVKGAKALYKFGRLQRGRYSPNGSDQAIF 440
H SDLA A AS+ FK + +YS L+ AK L+ F RG Y+ + + F
Sbjct: 172 HPGSDLAGETAAALAAASLAFKPFNSSYSALLLSHAKELFSFADKYRGLYTNSIPNAKAF 231
Query: 439 YNSTSYWDEFVWGGAWMYFATGNNSYLTVATAPGMAKHAGAYWFGSPNYGV--FTWDDKL 266
Y S+ Y DE +W AW++ ATG+ YL A + + G +GV F+WD+K
Sbjct: 232 YMSSGYSDELLWAAAWLHRATGDQYYLKYAM-------DNSGYMGGTGWGVKEFSWDNKY 284
Query: 265 PGAQVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPVFNSFNF--TKGGLIQLNHG 92
G Q+LLS++ L G Y L+ + + D C+ L +N T GGL+ +
Sbjct: 285 AGVQILLSKI-LLEGKGGIYTSTLKQYQTKADYFACACLKKNGGYNIQTTPGGLMYVREW 343
Query: 91 RPQPLQYAVNAAFLASLYSDYLEAAD 14
LQYA AA+L ++YSDYL AA+
Sbjct: 344 --NNLQYASAAAYLLAVYSDYLSAAN 367
>sptr|Q9FVQ2|Q9FVQ2 Endo-beta-1,4-glucanase, putative.
Length = 623
Score = 110 bits (274), Expect = 2e-23
Identities = 73/206 (35%), Positives = 105/206 (50%), Gaps = 5/206 (2%)
Frame = -2
Query: 616 HSCSDLAXXXXAXXXXASIVFKD-SKTYSDKLVKGAKALYKFGRLQRGRYSPNGSDQAIF 440
H SDLA A AS+ FK + +YS L+ AK L+ F RG Y+ + + F
Sbjct: 168 HPGSDLAGETAAALAAASLAFKPFNSSYSALLLSHAKELFSFADKYRGLYTNSIPNAKAF 227
Query: 439 YNSTSYWDEFVWGGAWMYFATGNNSYLTVATAPGMAKHAGAYWFGSPNYGV--FTWDDKL 266
Y S+ Y DE +W AW++ ATG+ YL A + + G +GV F+WD+K
Sbjct: 228 YMSSGYSDELLWAAAWLHRATGDQYYLKYAM-------DNSGYMGGTGWGVKEFSWDNKY 280
Query: 265 PGAQVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPVFNSFNF--TKGGLIQLNHG 92
G Q+LLS++ L G Y L+ + + D C+ L +N T GGL+ +
Sbjct: 281 AGVQILLSKI-LLEGKGGIYTSTLKQYQTKADYFACACLKKNGGYNIQTTPGGLMYVREW 339
Query: 91 RPQPLQYAVNAAFLASLYSDYLEAAD 14
LQYA AA+L ++YSDYL AA+
Sbjct: 340 --NNLQYASAAAYLLAVYSDYLSAAN 363
>sptr|Q9Z4I1|Q9Z4I1 Cellulase precursor (EC 3.2.1.4).
Length = 997
Score = 107 bits (267), Expect = 2e-22
Identities = 78/202 (38%), Positives = 103/202 (50%), Gaps = 8/202 (3%)
Frame = -2
Query: 607 SDLAXXXXAXXXXASIVFKDSK-TYSDKLVKGAKALYKFGRLQRGRYSPNGSDQAIFYNS 431
SDLA A +SIVF DS YS KL++ AK LY F RG+Y+ +D A FYNS
Sbjct: 185 SDLAAETAAALAASSIVFADSDPVYSAKLLQHAKELYNFADTYRGKYTDCITDAAAFYNS 244
Query: 430 -TSYWDEFVWGGAWMYFATGNNSYLTVATAPG---MAKHAGAYWFGSPNYGVFTWDDKLP 263
T Y DE WGGAW+Y AT +N+YL+ A + A W P WD K
Sbjct: 245 WTGYEDELAWGGAWLYLATNDNAYLSKALSAADRWSTSGGSANW---PYTWTQGWDSKHY 301
Query: 262 GAQVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPVFNS---FNFTKGGLIQLNHG 92
GAQ+LL+R+ L+ + I T N Y V + +T GGL L+
Sbjct: 302 GAQILLARITSNLNMPEATKFIQSTERN------LDYWTVGTNGGRVKYTPGGLAWLDQW 355
Query: 91 RPQPLQYAVNAAFLASLYSDYL 26
L+YA NAAF++ +YSD++
Sbjct: 356 --GSLRYAANAAFISFVYSDWV 375
>sw|P05522|GUN1_PERAE Endoglucanase 1 precursor (EC 3.2.1.4)
(Endo-1,4-beta-glucanase) (Abscission cellulase 1).
Length = 494
Score = 102 bits (255), Expect = 4e-21
Identities = 77/204 (37%), Positives = 108/204 (52%), Gaps = 7/204 (3%)
Frame = -2
Query: 607 SDLAXXXXAXXXXASIVFKDS-KTYSDKLVKGAKALYKFGRLQRGRYSPN-GSDQAIFYN 434
SD+A A ASIVF DS +YS KL+ A +++F RG YS + GS FY
Sbjct: 173 SDVAAETAAALAAASIVFGDSDSSYSTKLLHTAVKVFEFADQYRGSYSDSLGSVVCPFYC 232
Query: 433 STS-YWDEFVWGGAWMYFATGNNSYLTVATAPGMAKHAGAYWFGSPNYGVFTWDDKLPGA 257
S S Y DE +WG +W++ A+ N SY+T + G A +Y F+WDDK G
Sbjct: 233 SYSGYNDELLWGASWLHRASQNASYMTYIQSNGHTLGA-----DDDDYS-FSWDDKRVGT 286
Query: 256 QVLLSRLRLFLSPGYPYEEI--LRTFHNQTDNVMCSYLPVFNSF--NFTKGGLIQLNHGR 89
+VLLS+ G+ + I L+ + TDN +CS +P +SF +T GGL L G
Sbjct: 287 KVLLSK-------GFLQDRIEELQLYKVHTDNYICSLIPGTSSFQAQYTPGGL--LYKGS 337
Query: 88 PQPLQYAVNAAFLASLYSDYLEAA 17
LQY + AFL Y++YL ++
Sbjct: 338 ASNLQYVTSTAFLLLTYANYLNSS 361
>sptr|Q9S949|Q9S949 CEL3=CELLULASE 3 (Fragment).
Length = 176
Score = 101 bits (252), Expect = 8e-21
Identities = 49/84 (58%), Positives = 54/84 (64%)
Frame = -2
Query: 652 EXIDYXXXXXECHSCSDLAXXXXAXXXXASIVFKDSKTYSDKLVKGAKALYKFGRLQRGR 473
E IDY ECH CSDLA ASIVFKD+K YS KLV GA+ L+KF R QRGR
Sbjct: 93 EDIDYARPVTECHGCSDLAAEMARALASASIVFKDNKAYSQKLVHGARTLFKFSRDQRGR 152
Query: 472 YSPNGSDQAIFYNSTSYWDEFVWG 401
YS G++ FYNST YWDE WG
Sbjct: 153 YSV-GNEAETFYNSTGYWDELFWG 175
>sptr|Q9NCE3|Q9NCE3 Beta-1,4-endoglucanase 1.
Length = 450
Score = 99.4 bits (246), Expect = 4e-20
Identities = 57/131 (43%), Positives = 75/131 (57%), Gaps = 4/131 (3%)
Frame = -2
Query: 616 HSCSDLAXXXXAXXXXASIVFKD-SKTYSDKLVKGAKALYKFGRLQRGRYSPNGSDQAIF 440
H SDLA A ASIVFK TYS+ L+ AK LY F + RG+YS + +D A +
Sbjct: 164 HPGSDLAAETAAALAAASIVFKSVDPTYSNTLLTHAKQLYNFADMHRGKYSDSITDAAQY 223
Query: 439 YNSTSYWDEFVWGGAWMYFATGNNSYLTVATAPGMAKHAGAYW--FGSPNY-GVFTWDDK 269
Y+S Y D+ VWG W+Y AT +NSYLT A Y+ FG +Y G F+WD K
Sbjct: 224 YSSYGYEDDLVWGAVWLYKATNDNSYLT---------KAEQYYTQFGLADYDGYFSWDQK 274
Query: 268 LPGAQVLLSRL 236
+ G +VLL+ +
Sbjct: 275 ISGVEVLLAEI 285
>sptrnew|AAR29083|AAR29083 Cellulase (EC 3.2.1.4).
Length = 633
Score = 99.0 bits (245), Expect = 5e-20
Identities = 73/199 (36%), Positives = 96/199 (48%), Gaps = 4/199 (2%)
Frame = -2
Query: 607 SDLAXXXXAXXXXASIVFKDS-KTYSDKLVKGAKALYKFGRLQRGRYSPNGSDQAIFYNS 431
SDLA A ASI+FK + +YS+KL+ AK LY F RG+YS +D +YNS
Sbjct: 166 SDLAGGTAAALASASIIFKPTDSSYSEKLLAHAKQLYDFADRYRGKYSDCITDAQQYYNS 225
Query: 430 TS-YWDEFVWGGAWMYFATGNNSYLTVATAPGMAKHAGAYWFGSPNYGVFTWDDKLPGAQ 254
S Y DE WG W+Y AT YL A A A W P +WDD GAQ
Sbjct: 226 WSGYKDELTWGAVWLYLATEEQQYLDKALASVSDWGDPANW---PYRWTLSWDDVTYGAQ 282
Query: 253 VLLSRLRLFLSPGYPYEEILRTFHNQTD--NVMCSYLPVFNSFNFTKGGLIQLNHGRPQP 80
+LL+RL +++ D + S+ +T GGL L
Sbjct: 283 LLLARLT-------NDSRFVKSVERNLDYWSTGYSHNGSIERITYTPGGLAWLEQW--GS 333
Query: 79 LQYAVNAAFLASLYSDYLE 23
L+YA NAAFLA +YSD+++
Sbjct: 334 LRYASNAAFLAFVYSDWVD 352
>sptr|P94114|P94114 Endo-beta-1,4-glucanase (EC 3.2.1.4) (Cellulase)
(Endoglucanase) (Endo-1,4-beta-glucanase) (Carboxymethyl
cellulase).
Length = 497
Score = 99.0 bits (245), Expect = 5e-20
Identities = 75/199 (37%), Positives = 100/199 (50%), Gaps = 5/199 (2%)
Frame = -2
Query: 607 SDLAXXXXAXXXXASIVFKDSK-TYSDKLVKGAKALYKFGRLQRGRYSPN-GSDQAIFYN 434
SD+A A ASIVFKDS +YS KL+ A ++ F RG YS + GS FY
Sbjct: 176 SDVAAETAAALAAASIVFKDSDPSYSGKLLHTAMKVFDFADRYRGSYSDSIGSVVCPFYC 235
Query: 433 STS-YWDEFVWGGAWMYFATGNNSYLTVATAPGMAKHAGAYWFGSPNYGVFTWDDKLPGA 257
S S Y DE +WG +W++ A+ N+SYL + G A F F+WDDK PG
Sbjct: 236 SYSGYHDELLWGASWIHRASQNSSYLAYIKSNGHILGADDDGFS------FSWDDKRPGT 289
Query: 256 QVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPVFNSF--NFTKGGLIQLNHGRPQ 83
+VLLS + FL E + + +DN +CS LP ++F +T GGL L
Sbjct: 290 KVLLS--KNFLEKN---NEEFQLYKAHSDNYICSLLPGTSNFQAQYTPGGL--LYKASES 342
Query: 82 PLQYAVNAAFLASLYSDYL 26
LQY + L Y+ YL
Sbjct: 343 NLQYVTSTTLLLLTYAKYL 361
>sptr|Q9NCF1|Q9NCF1 Beta-1,4-glucanase 3 (Fragment).
Length = 270
Score = 98.6 bits (244), Expect = 7e-20
Identities = 61/129 (47%), Positives = 72/129 (55%), Gaps = 2/129 (1%)
Frame = -2
Query: 616 HSCSDLAXXXXAXXXXASIVFKDS-KTYSDKLVKGAKALYKFGRLQRGRYSPNGSDQAIF 440
H SDLA A ASIVFK S +YS L+ AK L+ F RG+YS + +D F
Sbjct: 108 HPGSDLAAETAAALAAASIVFKSSDSSYSKTLLTHAKQLFDFANKYRGKYSDSIADANNF 167
Query: 439 YNSTSYWDEFVWGGAWMYFATGNNSYLTVATAPGMAKHAG-AYWFGSPNYGVFTWDDKLP 263
Y S Y DE VWG AW+Y ATG++SYLT TA + K G YW G F WD K
Sbjct: 168 YASWDYKDELVWGAAWLYRATGDSSYLT--TAESLYKDFGIQYWNGD-----FGWDTKTS 220
Query: 262 GAQVLLSRL 236
G QVLL+ L
Sbjct: 221 GVQVLLADL 229
>sptr|Q9ZSP9|Q9ZSP9 Endo-beta-1,4-D-glucanase.
Length = 625
Score = 97.4 bits (241), Expect = 2e-19
Identities = 71/203 (34%), Positives = 101/203 (49%), Gaps = 3/203 (1%)
Frame = -2
Query: 616 HSCSDLAXXXXAXXXXASIVFKD-SKTYSDKLVKGAKALYKFGRLQRGRYSPNGSDQAIF 440
H SDLA A ASIVF+ + YS++L+ A L++F RG+Y + + +
Sbjct: 166 HPGSDLAGETAAAMAAASIVFRRYNPGYSNELLNHAHQLFEFADKYRGKYDSSITVAQKY 225
Query: 439 YNSTS-YWDEFVWGGAWMYFATGNNSYLTVATAPGMAKHAGAYWFGSPNYGVFTWDDKLP 263
Y S S Y DE +WG AW+Y A+ N YL G A G W + F WD K
Sbjct: 226 YRSVSGYADELLWGAAWLYKASNNQFYLNYLGRNGDAL-GGTGW----SMTEFGWDVKYA 280
Query: 262 GAQVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPVFN-SFNFTKGGLIQLNHGRP 86
G Q L+++ + G+ + + + +N MCS L N + T GGLI R
Sbjct: 281 GVQTLVAQFLMSGKAGH-NAPVFEKYQQKAENFMCSMLGKGNRNTQKTPGGLIY--RQRW 337
Query: 85 QPLQYAVNAAFLASLYSDYLEAA 17
+Q+ +AAFLA+ YSDYL +A
Sbjct: 338 NNMQFVTSAAFLATTYSDYLASA 360
>sptr|Q96546|Q96546 Endo-beta-1,4-glucanase (EC 3.2.1.4).
Length = 497
Score = 97.4 bits (241), Expect = 2e-19
Identities = 71/201 (35%), Positives = 105/201 (52%), Gaps = 5/201 (2%)
Frame = -2
Query: 607 SDLAXXXXAXXXXASIVFKDSK-TYSDKLVKGAKALYKFGRLQRGRYSPNGSDQAI-FYN 434
SD+A A ASIVFKDS +YS L++ A+ ++ F RG YS + S FY
Sbjct: 176 SDVAAETAAALAAASIVFKDSDPSYSSTLLRTAQKVFAFADKYRGSYSDSLSSVVCPFYC 235
Query: 433 STS-YWDEFVWGGAWMYFATGNNSYLTVATAPGMAKHAGAYWFGSPNYGVFTWDDKLPGA 257
S S Y DE +WG +W++ A+ + SYL+ + G A +Y F+WDDK PG
Sbjct: 236 SYSGYNDELLWGASWLHRASQDTSYLSYIQSNGQTMGA-----NDDDYS-FSWDDKRPGT 289
Query: 256 QVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPVFNSF--NFTKGGLIQLNHGRPQ 83
+++LS+ L S + + + +DN +CS +P SF +T GGL L G
Sbjct: 290 KIVLSKDFLEKS-----TQEFQAYKVHSDNYICSLIPGSPSFQAQYTPGGL--LFKGSES 342
Query: 82 PLQYAVNAAFLASLYSDYLEA 20
LQY +++FL Y+ YL +
Sbjct: 343 NLQYVTSSSFLLLTYAKYLRS 363
Database: /db/uniprot/tmp/swall
Posted date: Mar 5, 2004 7:45 PM
Number of letters in database: 442,889,342
Number of sequences in database: 1,395,590
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 491,820,200
Number of Sequences: 1395590
Number of extensions: 10628217
Number of successful extensions: 26666
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 25661
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26327
length of database: 442,889,342
effective HSP length: 119
effective length of database: 276,814,132
effective search space used: 30449554520
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

