BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= QAF25f04.yg.3.3
(1599 letters)
Database: swall
1,381,838 sequences; 439,479,560 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
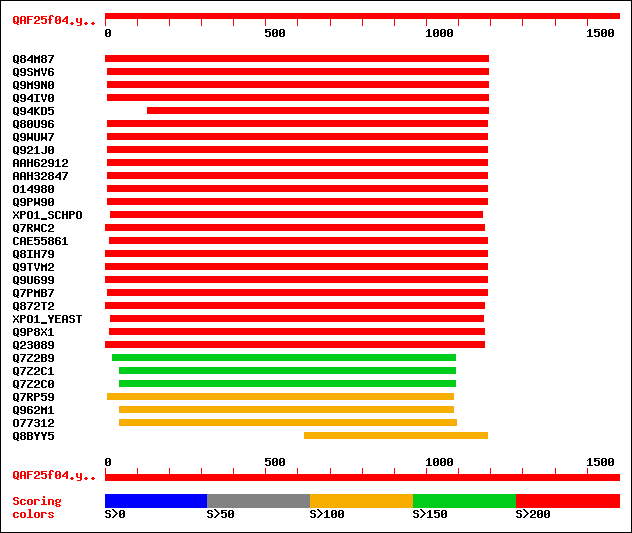
sptr|Q84M87|Q84M87 Putative chromosome region maintenance protein. 731 0.0
sptr|Q9SMV6|Q9SMV6 EXPORTIN1 (XPO1) protein (XPO1) (Putative EXP... 606 e-172
sptr|Q9M9N0|Q9M9N0 Putative exportin1 (XPO1) protein. 589 e-167
sptr|Q94IV0|Q94IV0 Exportin 1b. 586 e-166
sptr|Q94KD5|Q94KD5 AT3g03110/T17B22_20. 533 e-150
sptr|Q80U96|Q80U96 Nuclear export factor CRM1. 339 6e-92
sptr|Q9WUW7|Q9WUW7 CRM1 protein (Fragment). 339 6e-92
sptr|Q921J0|Q921J0 Similar to exportin 1 (CRM1, yeast, homolog) ... 338 1e-91
sptrnew|AAH62912|AAH62912 Xpo1 protein. 338 1e-91
sptrnew|AAH32847|AAH32847 Exportin 1. 335 2e-90
sptr|O14980|O14980 CRM1 protein. 335 2e-90
sptr|Q9PW90|Q9PW90 CRM1/XPO1 protein. 329 7e-89
sw|P14068|XPO1_SCHPO Exportin 1 (Chromosome region maintenance p... 313 4e-84
sptr|Q7RWC2|Q7RWC2 Hypothetical protein. 307 3e-82
sptrnew|CAE55861|CAE55861 Exportin 1. 301 1e-80
sptr|Q8IH79|Q8IH79 GH01059p (Fragment). 295 1e-78
sptr|Q9TVM2|Q9TVM2 Chromosomal region maintenance 1 protein (LD4... 295 1e-78
sptr|Q9U699|Q9U699 Chromosomal region maintenance protein CRM1. 295 1e-78
sptr|Q7PMB7|Q7PMB7 ENSANGP00000013197 (Fragment). 291 2e-77
sptr|Q872T2|Q872T2 Probable nuclear export factor CRM1 (Fragment). 288 1e-76
sw|P30822|XPO1_YEAST Exportin 1 (Chromosome region maintenance p... 274 3e-72
sptr|Q9P8X1|Q9P8X1 Crm1p. 260 5e-68
sptr|Q23089|Q23089 Hypothetical protein. 255 1e-66
sptr|Q7Z2B9|Q7Z2B9 CRM1 (Fragment). 167 4e-40
sptr|Q7Z2C1|Q7Z2C1 Exportin 1. 153 8e-36
sptr|Q7Z2C0|Q7Z2C0 CRM1. 153 8e-36
sptr|Q7RP59|Q7RP59 Plasmodium vivax PV1H14065_P-related. 149 1e-34
sptr|Q962M1|Q962M1 PV1H14065_P. 149 1e-34
sptr|O77312|O77312 PFC0135c, MAL3P1.9 protein. 147 6e-34
sptr|Q8BYY5|Q8BYY5 CRM1 protein (Fragment). 145 2e-33
>sptr|Q84M87|Q84M87 Putative chromosome region maintenance protein.
Length = 1070
Score = 731 bits (1888), Expect = 0.0
Identities = 372/398 (93%), Positives = 375/398 (94%)
Frame = +1
Query: 1 SLSIDILKNQDVIRSVLNILQTNTSVASSLGPHFFPQISLIFLDMLTVYRMYSELVSSTI 180
S SIDILKNQDVIRSVLNILQTNTSVASSLGPHFFPQISLIFLDMLTVYRMYSELVSSTI
Sbjct: 673 SQSIDILKNQDVIRSVLNILQTNTSVASSLGPHFFPQISLIFLDMLTVYRMYSELVSSTI 732
Query: 181 AEGGPFASRTSFVKLLRSVKRETLKLIETFVDKAEDLPHVGKQFVPPMMDPVLGDYARNV 360
AEGGPFASRTSFVKLLRSVKRETLKLIETFVDKAEDLPH+GKQFVPPMMDP+L DYARNV
Sbjct: 733 AEGGPFASRTSFVKLLRSVKRETLKLIETFVDKAEDLPHIGKQFVPPMMDPILADYARNV 792
Query: 361 PDARESEVLSLFATIINKYKGEMLEDVPRIFEAVFQCTLEMITKNFEDYPEHRLKFFSLL 540
PDARESEVLSLFATIINKYKGEMLEDVPRIFEAVFQCTLEMITKNFEDYPEHRLKFFSLL
Sbjct: 793 PDARESEVLSLFATIINKYKGEMLEDVPRIFEAVFQCTLEMITKNFEDYPEHRLKFFSLL 852
Query: 541 RAIGTHCFQALIQLSSQQLKLVIDSINWAFRHTERNIAETGLSLLLEILKKFQASGFQNQ 720
RAIGTHCFQALIQLSSQQLKLVIDSINWAFRHTERNIAETGLSLLLEILK FQASGFQNQ
Sbjct: 853 RAIGTHCFQALIQLSSQQLKLVIDSINWAFRHTERNIAETGLSLLLEILKNFQASGFQNQ 912
Query: 721 FYKTYFLTIEQEIFAVLTDTFHKPGFKLHVSVLQHLFCAVDGLTEPLWDASSVPYQYTDN 900
FYKTYFL IEQEIFAVLTDTFHKPGFKLHV VLQHLFC VDGLTEPLWDASSVPYQYTDN
Sbjct: 913 FYKTYFLNIEQEIFAVLTDTFHKPGFKLHVLVLQHLFCVVDGLTEPLWDASSVPYQYTDN 972
Query: 901 AMFVRDYTIKLLGTSFPNMTVAEVTKFVDGLLSSKLDLPSFKNHIRDFLVQSKEFSAQDN 1080
AMFVRDYTIKLLG+SFPNMT EVTKFVDGLLSSK DLPSFKNHIRDFLVQSKEFSAQDN
Sbjct: 973 AMFVRDYTIKLLGSSFPNMTPTEVTKFVDGLLSSKHDLPSFKNHIRDFLVQSKEFSAQDN 1032
Query: 1081 KDLYXXXXXXXXXXXXXXMLAIPGLIAPSELQDEMVDS 1194
KDLY MLAIPGLIAPSELQDEMVDS
Sbjct: 1033 KDLYAEEAAAQRERERQRMLAIPGLIAPSELQDEMVDS 1070
>sptr|Q9SMV6|Q9SMV6 EXPORTIN1 (XPO1) protein (XPO1) (Putative
EXPORTIN1 (XPO1) protein).
Length = 1075
Score = 606 bits (1563), Expect = e-172
Identities = 309/398 (77%), Positives = 341/398 (85%), Gaps = 2/398 (0%)
Frame = +1
Query: 7 SIDILKNQDVIRSVLNILQTNTSVASSLGPHFFPQISLIFLDMLTVYRMYSELVSSTIAE 186
S++ LK+Q VIR+VLNILQTNTS A+SLG +F QISLIFLDML VYRMYSELVS+ I E
Sbjct: 678 SVEFLKDQVVIRTVLNILQTNTSAATSLGTYFLSQISLIFLDMLNVYRMYSELVSTNITE 737
Query: 187 GGPFASRTSFVKLLRSVKRETLKLIETFVDKAEDLPHVGKQFVPPMMDPVLGDYARNVPD 366
GGP+AS+TSFVKLLRSVKRETLKLIETF+DKAED PH+GKQFVPPMM+ VLGDYARNVPD
Sbjct: 738 GGPYASKTSFVKLLRSVKRETLKLIETFLDKAEDQPHIGKQFVPPMMESVLGDYARNVPD 797
Query: 367 ARESEVLSLFATIINKYKGEMLEDVPRIFEAVFQCTLEMITKNFEDYPEHRLKFFSLLRA 546
ARESEVLSLFATIINKYK ML+DVP IFEAVFQCTLEMITKNFEDYPEHRLKFFSLLRA
Sbjct: 798 ARESEVLSLFATIINKYKATMLDDVPHIFEAVFQCTLEMITKNFEDYPEHRLKFFSLLRA 857
Query: 547 IGTHCFQALIQLSSQQLKLVIDSINWAFRHTERNIAETGLSLLLEILKKFQASGFQNQFY 726
I T CF ALI+LSS QLKLV+DSI WAFRHTERNIAETGL+LLLE+LK FQ S F NQFY
Sbjct: 858 IATFCFPALIKLSSPQLKLVMDSIIWAFRHTERNIAETGLNLLLEMLKNFQQSEFCNQFY 917
Query: 727 KTYFLTIEQEIFAVLTDTFHKPGFKLHVSVLQHLFCAVD--GLTEPLWDASSVPYQYTDN 900
++YF+ IEQEIFAVLTDTFHKPGFKLHV VLQ LFC + LTEPLWDA++VPY Y DN
Sbjct: 918 RSYFMQIEQEIFAVLTDTFHKPGFKLHVLVLQQLFCLPESGALTEPLWDATTVPYPYPDN 977
Query: 901 AMFVRDYTIKLLGTSFPNMTVAEVTKFVDGLLSSKLDLPSFKNHIRDFLVQSKEFSAQDN 1080
FVR+YTIKLL +SFPNMT AEVT+FV+GL S+ D FKN+IRDFLVQSKEFSAQDN
Sbjct: 978 VAFVREYTIKLLSSSFPNMTAAEVTQFVNGLYESRNDPSGFKNNIRDFLVQSKEFSAQDN 1037
Query: 1081 KDLYXXXXXXXXXXXXXXMLAIPGLIAPSELQDEMVDS 1194
KDLY ML+IPGLIAP+E+QDEMVDS
Sbjct: 1038 KDLYAEEAAAQRERERQRMLSIPGLIAPNEIQDEMVDS 1075
>sptr|Q9M9N0|Q9M9N0 Putative exportin1 (XPO1) protein.
Length = 1022
Score = 589 bits (1518), Expect = e-167
Identities = 301/398 (75%), Positives = 337/398 (84%), Gaps = 2/398 (0%)
Frame = +1
Query: 7 SIDILKNQDVIRSVLNILQTNTSVASSLGPHFFPQISLIFLDMLTVYRMYSELVSSTIAE 186
S DILK DVIR+VLNILQTNT VA+SLG F QISLIFLDML VYRMYSELVSS+IA
Sbjct: 625 SADILKEPDVIRTVLNILQTNTRVATSLGTFFLSQISLIFLDMLNVYRMYSELVSSSIAN 684
Query: 187 GGPFASRTSFVKLLRSVKRETLKLIETFVDKAEDLPHVGKQFVPPMMDPVLGDYARNVPD 366
GGP+ASRTS VKLLRSVKRE LKLIETF+DKAE+ PH+GKQFVPPMMD VLGDYARNVPD
Sbjct: 685 GGPYASRTSLVKLLRSVKREILKLIETFLDKAENQPHIGKQFVPPMMDQVLGDYARNVPD 744
Query: 367 ARESEVLSLFATIINKYKGEMLEDVPRIFEAVFQCTLEMITKNFEDYPEHRLKFFSLLRA 546
ARESEVLSLFATIINKYK M ++VP IFEAVFQCTLEMITKNFEDYPEHRLKFFSLLRA
Sbjct: 745 ARESEVLSLFATIINKYKVVMRDEVPLIFEAVFQCTLEMITKNFEDYPEHRLKFFSLLRA 804
Query: 547 IGTHCFQALIQLSSQQLKLVIDSINWAFRHTERNIAETGLSLLLEILKKFQASGFQNQFY 726
I T CF+ALIQLSS+QLKLV+DS+ WAFRHTERNIAETGL+LLLE+LK FQ S F N+FY
Sbjct: 805 IATFCFRALIQLSSEQLKLVMDSVIWAFRHTERNIAETGLNLLLEMLKNFQKSDFCNKFY 864
Query: 727 KTYFLTIEQEIFAVLTDTFHKPGFKLHVSVLQHLFCAVD--GLTEPLWDASSVPYQYTDN 900
+TYFL IEQE+FAVLTDTFHKPGFKLHV VLQHLF V+ L EPLWDA++VP+ Y++N
Sbjct: 865 QTYFLQIEQEVFAVLTDTFHKPGFKLHVLVLQHLFSLVESGSLAEPLWDAATVPHPYSNN 924
Query: 901 AMFVRDYTIKLLGTSFPNMTVAEVTKFVDGLLSSKLDLPSFKNHIRDFLVQSKEFSAQDN 1080
FV +YT KLL +SFPNMT EVT+FV+GL S+ D+ FK++IRDFL+QSKEFSAQDN
Sbjct: 925 VAFVLEYTTKLLSSSFPNMTTTEVTQFVNGLYESRNDVGRFKDNIRDFLIQSKEFSAQDN 984
Query: 1081 KDLYXXXXXXXXXXXXXXMLAIPGLIAPSELQDEMVDS 1194
KDLY ML+IPGLIAPSE+QD+M DS
Sbjct: 985 KDLYAEEAAAQMERERQRMLSIPGLIAPSEIQDDMADS 1022
>sptr|Q94IV0|Q94IV0 Exportin 1b.
Length = 1076
Score = 586 bits (1510), Expect = e-166
Identities = 300/398 (75%), Positives = 336/398 (84%), Gaps = 2/398 (0%)
Frame = +1
Query: 7 SIDILKNQDVIRSVLNILQTNTSVASSLGPHFFPQISLIFLDMLTVYRMYSELVSSTIAE 186
S DILK DVIR+VLNILQTNT VA+SLG F QISLIFLDML VYRMYSELVSS+IA
Sbjct: 679 SADILKEPDVIRTVLNILQTNTRVATSLGTFFLSQISLIFLDMLNVYRMYSELVSSSIAN 738
Query: 187 GGPFASRTSFVKLLRSVKRETLKLIETFVDKAEDLPHVGKQFVPPMMDPVLGDYARNVPD 366
GGP+ASRTS VKLLRSVKRE LKLIETF+DKAE+ PH+GKQFVPPMMD VLGDYARNVPD
Sbjct: 739 GGPYASRTSLVKLLRSVKREILKLIETFLDKAENQPHIGKQFVPPMMDQVLGDYARNVPD 798
Query: 367 ARESEVLSLFATIINKYKGEMLEDVPRIFEAVFQCTLEMITKNFEDYPEHRLKFFSLLRA 546
ARESEVLSLFATIINKYK M ++VP IFEAVFQCTLEMITKNFEDYPEHRLKFFSLLRA
Sbjct: 799 ARESEVLSLFATIINKYKVVMRDEVPLIFEAVFQCTLEMITKNFEDYPEHRLKFFSLLRA 858
Query: 547 IGTHCFQALIQLSSQQLKLVIDSINWAFRHTERNIAETGLSLLLEILKKFQASGFQNQFY 726
I T CF+ALIQLSS+QLKLV+DS+ WAFRHTERNIAETGL+LLLE+LK FQ S F N+FY
Sbjct: 859 IATFCFRALIQLSSEQLKLVMDSVIWAFRHTERNIAETGLNLLLEMLKNFQKSDFCNKFY 918
Query: 727 KTYFLTIEQEIFAVLTDTFHKPGFKLHVSVLQHLFCAVD--GLTEPLWDASSVPYQYTDN 900
+TYFL IEQE+FAVLTDTFHK GFKLHV VLQHLF V+ L EPLWDA++VP+ Y++N
Sbjct: 919 QTYFLQIEQEVFAVLTDTFHKSGFKLHVLVLQHLFSLVESGSLAEPLWDAATVPHPYSNN 978
Query: 901 AMFVRDYTIKLLGTSFPNMTVAEVTKFVDGLLSSKLDLPSFKNHIRDFLVQSKEFSAQDN 1080
FV +YT KLL +SFPNMT EVT+FV+GL S+ D+ FK++IRDFL+QSKEFSAQDN
Sbjct: 979 VAFVLEYTTKLLSSSFPNMTTTEVTQFVNGLYESRNDVGRFKDNIRDFLIQSKEFSAQDN 1038
Query: 1081 KDLYXXXXXXXXXXXXXXMLAIPGLIAPSELQDEMVDS 1194
KDLY ML+IPGLIAPSE+QD+M DS
Sbjct: 1039 KDLYAEEAAAQMERERQRMLSIPGLIAPSEIQDDMADS 1076
>sptr|Q94KD5|Q94KD5 AT3g03110/T17B22_20.
Length = 356
Score = 533 bits (1372), Expect = e-150
Identities = 269/356 (75%), Positives = 303/356 (85%), Gaps = 2/356 (0%)
Frame = +1
Query: 133 MLTVYRMYSELVSSTIAEGGPFASRTSFVKLLRSVKRETLKLIETFVDKAEDLPHVGKQF 312
ML VYRMYSELVSS+IA GGP+ASRTS VKLLRSVKRE LKLIETF+DKAE+ PH+GKQF
Sbjct: 1 MLNVYRMYSELVSSSIANGGPYASRTSLVKLLRSVKREILKLIETFLDKAENQPHIGKQF 60
Query: 313 VPPMMDPVLGDYARNVPDARESEVLSLFATIINKYKGEMLEDVPRIFEAVFQCTLEMITK 492
VPPMMD VLGDYARNVPDARESEVLSLFATIINKYK M ++VP IFEAVFQCTLEMITK
Sbjct: 61 VPPMMDQVLGDYARNVPDARESEVLSLFATIINKYKVVMRDEVPLIFEAVFQCTLEMITK 120
Query: 493 NFEDYPEHRLKFFSLLRAIGTHCFQALIQLSSQQLKLVIDSINWAFRHTERNIAETGLSL 672
NFEDYPEHRLKFFSLLRAI T CF+ALIQLSS+QLKLV+DS+ WAFRHTERNIAETGL+L
Sbjct: 121 NFEDYPEHRLKFFSLLRAIATFCFRALIQLSSEQLKLVMDSVIWAFRHTERNIAETGLNL 180
Query: 673 LLEILKKFQASGFQNQFYKTYFLTIEQEIFAVLTDTFHKPGFKLHVSVLQHLFCAVD--G 846
LLE+LK FQ S F N+FY+TYFL IEQE+FAVLTDTFHKPGFKLHV VLQHLF V+
Sbjct: 181 LLEMLKNFQKSDFCNKFYQTYFLQIEQEVFAVLTDTFHKPGFKLHVLVLQHLFSLVESGS 240
Query: 847 LTEPLWDASSVPYQYTDNAMFVRDYTIKLLGTSFPNMTVAEVTKFVDGLLSSKLDLPSFK 1026
L EPLWDA++VP+ Y++N FV +YT KLL +SFPNMT EVT+FV+GL S+ D+ FK
Sbjct: 241 LAEPLWDAATVPHPYSNNVAFVLEYTTKLLSSSFPNMTTTEVTQFVNGLYESRNDVGRFK 300
Query: 1027 NHIRDFLVQSKEFSAQDNKDLYXXXXXXXXXXXXXXMLAIPGLIAPSELQDEMVDS 1194
++IRDFL+QSKEFSAQDNKDLY ML+IPGLIAPSE+QD+M DS
Sbjct: 301 DNIRDFLIQSKEFSAQDNKDLYAEEAAAQMERERQRMLSIPGLIAPSEIQDDMADS 356
>sptr|Q80U96|Q80U96 Nuclear export factor CRM1.
Length = 1071
Score = 339 bits (870), Expect = 6e-92
Identities = 174/402 (43%), Positives = 253/402 (62%), Gaps = 7/402 (1%)
Frame = +1
Query: 7 SIDILKNQDVIRSVLNILQTNTSVASSLGPHFFPQISLIFLDMLTVYRMYSELVSSTIAE 186
++DILK+ + ++ + +IL+TN ++G F Q+ I+LDML VY+ SE +S+ I
Sbjct: 675 NVDILKDPETVKQLGSILKTNVRACKAVGHPFVIQLGRIYLDMLNVYKCLSENISAAIQA 734
Query: 187 GGPFASRTSFVKLLRSVKRETLKLIETFVDKAEDLPHVGKQFVPPMMDPVLGDYARNVPD 366
G ++ ++ +R+VKRETLKLI +V ++ D V + FVPP++D VL DY RNVP
Sbjct: 735 NGEMVTKQPLIRSMRTVKRETLKLISGWVSRSNDPQMVAENFVPPLLDAVLIDYQRNVPA 794
Query: 367 ARESEVLSLFATIINKYKGEMLEDVPRIFEAVFQCTLEMITKNFEDYPEHRLKFFSLLRA 546
ARE EVLS A I+NK G + ++P+IF+AVF+CTL MI K+FE+YPEHR FF LL+A
Sbjct: 795 AREPEVLSTMAIIVNKLGGHITAEIPQIFDAVFECTLNMINKDFEEYPEHRTNFFLLLQA 854
Query: 547 IGTHCFQALIQLSSQQLKLVIDSINWAFRHTERNIAETGLSLLLEILKKF-QASGFQNQF 723
+ +HCF A + + Q KLV+DSI WAF+HT RN+A+TGL +L +L+ Q F
Sbjct: 855 VNSHCFPAFLAIPPAQFKLVLDSIIWAFKHTMRNVADTGLQILFTLLQNVAQEEAAAQSF 914
Query: 724 YKTYFLTIEQEIFAVLTDTFHKPGFKLHVSVLQHLFCAVD--GLTEPLWDASSVPYQYTD 897
Y+TYF I Q IF+V+TDT H G +H S+L ++F V+ ++ PL S V
Sbjct: 915 YQTYFCDILQHIFSVVTDTSHTAGLTMHASILAYMFNLVEEGKISTPLNPGSPV-----S 969
Query: 898 NAMFVRDYTIKLLGTSFPNMTVAEVTKFVDGLLSSKLDLPSFKNHIRDFLVQSKEFSAQD 1077
N MF++DY LL ++FP++ A+V FV GL S D+P+FK H+RDFLVQ KEF+ +D
Sbjct: 970 NQMFIQDYVANLLKSAFPHLQDAQVKLFVTGLFSLNQDIPAFKEHLRDFLVQIKEFAGED 1029
Query: 1078 NKDLY----XXXXXXXXXXXXXXMLAIPGLIAPSELQDEMVD 1191
DL+ +++PG++ P E+ +EM D
Sbjct: 1030 TSDLFLEERETALRQAQEEKHKLQMSVPGILNPHEIPEEMCD 1071
>sptr|Q9WUW7|Q9WUW7 CRM1 protein (Fragment).
Length = 625
Score = 339 bits (870), Expect = 6e-92
Identities = 174/402 (43%), Positives = 253/402 (62%), Gaps = 7/402 (1%)
Frame = +1
Query: 7 SIDILKNQDVIRSVLNILQTNTSVASSLGPHFFPQISLIFLDMLTVYRMYSELVSSTIAE 186
++DILK+ + ++ + +IL+TN ++G F Q+ I+LDML VY+ SE +S+ I
Sbjct: 229 NVDILKDPETVKQLGSILKTNVRACKAVGHPFVIQLGRIYLDMLNVYKCLSENISAAIQA 288
Query: 187 GGPFASRTSFVKLLRSVKRETLKLIETFVDKAEDLPHVGKQFVPPMMDPVLGDYARNVPD 366
G ++ ++ +R+VKRETLKLI +V ++ D V + FVPP++D VL DY RNVP
Sbjct: 289 NGEMVTKQPLIRSMRTVKRETLKLISGWVSRSNDPQMVAENFVPPLLDAVLIDYQRNVPA 348
Query: 367 ARESEVLSLFATIINKYKGEMLEDVPRIFEAVFQCTLEMITKNFEDYPEHRLKFFSLLRA 546
ARE EVLS A I+NK G + ++P+IF+AVF+CTL MI K+FE+YPEHR FF LL+A
Sbjct: 349 AREPEVLSTMAIIVNKLGGHITAEIPQIFDAVFECTLNMINKDFEEYPEHRTNFFLLLQA 408
Query: 547 IGTHCFQALIQLSSQQLKLVIDSINWAFRHTERNIAETGLSLLLEILKKF-QASGFQNQF 723
+ +HCF A + + Q KLV+DSI WAF+HT RN+A+TGL +L +L+ Q F
Sbjct: 409 VNSHCFPAFLAIPPAQFKLVLDSIIWAFKHTMRNVADTGLQILFTLLQNVAQEEAAAQSF 468
Query: 724 YKTYFLTIEQEIFAVLTDTFHKPGFKLHVSVLQHLFCAVD--GLTEPLWDASSVPYQYTD 897
Y+TYF I Q IF+V+TDT H G +H S+L ++F V+ ++ PL S V
Sbjct: 469 YQTYFCDILQHIFSVVTDTSHTAGLTMHASILAYMFNLVEEGKISTPLNPGSPV-----S 523
Query: 898 NAMFVRDYTIKLLGTSFPNMTVAEVTKFVDGLLSSKLDLPSFKNHIRDFLVQSKEFSAQD 1077
N MF++DY LL ++FP++ A+V FV GL S D+P+FK H+RDFLVQ KEF+ +D
Sbjct: 524 NQMFIQDYVANLLKSAFPHLQDAQVKLFVTGLFSLNQDIPAFKEHLRDFLVQIKEFAGED 583
Query: 1078 NKDLY----XXXXXXXXXXXXXXMLAIPGLIAPSELQDEMVD 1191
DL+ +++PG++ P E+ +EM D
Sbjct: 584 TSDLFLEERETALRQAQEEKHKLQMSVPGILNPHEIPEEMCD 625
>sptr|Q921J0|Q921J0 Similar to exportin 1 (CRM1, yeast, homolog)
(Expressed sequence AA420417).
Length = 564
Score = 338 bits (868), Expect = 1e-91
Identities = 173/402 (43%), Positives = 254/402 (63%), Gaps = 7/402 (1%)
Frame = +1
Query: 7 SIDILKNQDVIRSVLNILQTNTSVASSLGPHFFPQISLIFLDMLTVYRMYSELVSSTIAE 186
++DILK+ + ++ + +IL+TN ++G F Q+ I+LDML VY+ SE +S+ I
Sbjct: 168 NVDILKDPETVKQLGSILKTNVRACKAVGHPFVIQLGRIYLDMLNVYKCLSENISAAIQA 227
Query: 187 GGPFASRTSFVKLLRSVKRETLKLIETFVDKAEDLPHVGKQFVPPMMDPVLGDYARNVPD 366
G ++ ++ +R+VKRETLKLI +V ++ D V + FVPP++D VL DY RNVP
Sbjct: 228 NGEMVTKQPLIRSMRTVKRETLKLISGWVSRSNDPQMVAENFVPPLLDAVLIDYQRNVPA 287
Query: 367 ARESEVLSLFATIINKYKGEMLEDVPRIFEAVFQCTLEMITKNFEDYPEHRLKFFSLLRA 546
ARE EVLS A I+NK G + ++P+IF+AVF+CTL MI K+FE+YPEHR FF LL+A
Sbjct: 288 AREPEVLSTMAIIVNKLGGHITAEIPQIFDAVFECTLNMINKDFEEYPEHRTNFFLLLQA 347
Query: 547 IGTHCFQALIQLSSQQLKLVIDSINWAFRHTERNIAETGLSLLLEILKKF-QASGFQNQF 723
+ +HCF A + + Q KLV+DSI WAF+HT RN+A+TGL +L +L+ Q F
Sbjct: 348 VNSHCFPAFLAIPPAQFKLVLDSIIWAFKHTMRNVADTGLQILFTLLQNVAQEEAAAQSF 407
Query: 724 YKTYFLTIEQEIFAVLTDTFHKPGFKLHVSVLQHLFCAVD--GLTEPLWDASSVPYQYTD 897
Y+TYF I Q IF+V+TDT H G +H S+L ++F V+ ++ PL + V +
Sbjct: 408 YQTYFCDILQHIFSVVTDTSHTAGLTMHASILAYMFNLVEEGKISTPLNPGNPV-----N 462
Query: 898 NAMFVRDYTIKLLGTSFPNMTVAEVTKFVDGLLSSKLDLPSFKNHIRDFLVQSKEFSAQD 1077
N MF++DY LL ++FP++ A+V FV GL S D+P+FK H+RDFLVQ KEF+ +D
Sbjct: 463 NQMFIQDYVANLLKSAFPHLQDAQVKLFVTGLFSLNQDIPAFKEHLRDFLVQIKEFAGED 522
Query: 1078 NKDLY----XXXXXXXXXXXXXXMLAIPGLIAPSELQDEMVD 1191
DL+ +++PG++ P E+ +EM D
Sbjct: 523 TSDLFLEERETALRQAQEEKHKLQMSVPGILNPHEIPEEMCD 564
>sptrnew|AAH62912|AAH62912 Xpo1 protein.
Length = 1071
Score = 338 bits (868), Expect = 1e-91
Identities = 173/402 (43%), Positives = 254/402 (63%), Gaps = 7/402 (1%)
Frame = +1
Query: 7 SIDILKNQDVIRSVLNILQTNTSVASSLGPHFFPQISLIFLDMLTVYRMYSELVSSTIAE 186
++DILK+ + ++ + +IL+TN ++G F Q+ I+LDML VY+ SE +S+ I
Sbjct: 675 NVDILKDPETVKQLGSILKTNVRACKAVGHPFVIQLGRIYLDMLNVYKCLSENISAAIQA 734
Query: 187 GGPFASRTSFVKLLRSVKRETLKLIETFVDKAEDLPHVGKQFVPPMMDPVLGDYARNVPD 366
G ++ ++ +R+VKRETLKLI +V ++ D V + FVPP++D VL DY RNVP
Sbjct: 735 NGEMVTKQPLIRSMRTVKRETLKLISGWVSRSNDPQMVAENFVPPLLDAVLIDYQRNVPA 794
Query: 367 ARESEVLSLFATIINKYKGEMLEDVPRIFEAVFQCTLEMITKNFEDYPEHRLKFFSLLRA 546
ARE EVLS A I+NK G + ++P+IF+AVF+CTL MI K+FE+YPEHR FF LL+A
Sbjct: 795 AREPEVLSTMAIIVNKLGGHITAEIPQIFDAVFECTLNMINKDFEEYPEHRTNFFLLLQA 854
Query: 547 IGTHCFQALIQLSSQQLKLVIDSINWAFRHTERNIAETGLSLLLEILKKF-QASGFQNQF 723
+ +HCF A + + Q KLV+DSI WAF+HT RN+A+TGL +L +L+ Q F
Sbjct: 855 VNSHCFPAFLAIPPAQFKLVLDSIIWAFKHTMRNVADTGLQILFTLLQNVAQEEAAAQSF 914
Query: 724 YKTYFLTIEQEIFAVLTDTFHKPGFKLHVSVLQHLFCAVD--GLTEPLWDASSVPYQYTD 897
Y+TYF I Q IF+V+TDT H G +H S+L ++F V+ ++ PL + V +
Sbjct: 915 YQTYFCDILQHIFSVVTDTSHTAGLTMHASILAYMFNLVEEGKISTPLNPGNPV-----N 969
Query: 898 NAMFVRDYTIKLLGTSFPNMTVAEVTKFVDGLLSSKLDLPSFKNHIRDFLVQSKEFSAQD 1077
N MF++DY LL ++FP++ A+V FV GL S D+P+FK H+RDFLVQ KEF+ +D
Sbjct: 970 NQMFIQDYVANLLKSAFPHLQDAQVKLFVTGLFSLNQDIPAFKEHLRDFLVQIKEFAGED 1029
Query: 1078 NKDLY----XXXXXXXXXXXXXXMLAIPGLIAPSELQDEMVD 1191
DL+ +++PG++ P E+ +EM D
Sbjct: 1030 TSDLFLEERETALRQAQEEKHKLQMSVPGILNPHEIPEEMCD 1071
>sptrnew|AAH32847|AAH32847 Exportin 1.
Length = 1071
Score = 335 bits (858), Expect = 2e-90
Identities = 171/400 (42%), Positives = 250/400 (62%), Gaps = 5/400 (1%)
Frame = +1
Query: 7 SIDILKNQDVIRSVLNILQTNTSVASSLGPHFFPQISLIFLDMLTVYRMYSELVSSTIAE 186
++DILK+ + ++ + +IL+TN ++G F Q+ I+LDML VY+ SE +S+ I
Sbjct: 675 NVDILKDPETVKQLGSILKTNVRACKAVGHPFVIQLGRIYLDMLNVYKCLSENISAAIQA 734
Query: 187 GGPFASRTSFVKLLRSVKRETLKLIETFVDKAEDLPHVGKQFVPPMMDPVLGDYARNVPD 366
G ++ ++ +R+VKRETLKLI +V ++ D V + FVPP++D VL DY RNVP
Sbjct: 735 NGEMVTKQPLIRSMRTVKRETLKLISGWVSRSNDPQMVAENFVPPLLDAVLIDYQRNVPA 794
Query: 367 ARESEVLSLFATIINKYKGEMLEDVPRIFEAVFQCTLEMITKNFEDYPEHRLKFFSLLRA 546
ARE EVLS A I+NK G + ++P+IF+AVF+CTL MI K+FE+YPEHR FF LL+A
Sbjct: 795 AREPEVLSTMAIIVNKLGGHITAEIPQIFDAVFECTLNMINKDFEEYPEHRTNFFLLLQA 854
Query: 547 IGTHCFQALIQLSSQQLKLVIDSINWAFRHTERNIAETGLSLLLEILKKF-QASGFQNQF 723
+ +HCF A + + Q KLV+DSI WAF+HT RN+A+TGL +L +L+ Q F
Sbjct: 855 VNSHCFPAFLAIPPTQFKLVLDSIIWAFKHTMRNVADTGLQILFTLLQNVAQEEAAAQSF 914
Query: 724 YKTYFLTIEQEIFAVLTDTFHKPGFKLHVSVLQHLFCAVDGLTEPLWDASSVPYQYTDNA 903
Y+TYF I Q IF+V+TDT H G +H S+L ++F V+ E S P +N
Sbjct: 915 YQTYFCDILQHIFSVVTDTSHTAGLTMHASILAYMFNLVE---EGKISTSLNPGNPVNNQ 971
Query: 904 MFVRDYTIKLLGTSFPNMTVAEVTKFVDGLLSSKLDLPSFKNHIRDFLVQSKEFSAQDNK 1083
+F+++Y LL ++FP++ A+V FV GL S D+P+FK H+RDFLVQ KEF+ +D
Sbjct: 972 IFLQEYVANLLKSAFPHLQDAQVKLFVTGLFSLNQDIPAFKEHLRDFLVQIKEFAGEDTS 1031
Query: 1084 DLY----XXXXXXXXXXXXXXMLAIPGLIAPSELQDEMVD 1191
DL+ +++PG+ P E+ +EM D
Sbjct: 1032 DLFLEEREIALRQADEEKHKRQMSVPGIFNPHEIPEEMCD 1071
>sptr|O14980|O14980 CRM1 protein.
Length = 1071
Score = 335 bits (858), Expect = 2e-90
Identities = 171/400 (42%), Positives = 250/400 (62%), Gaps = 5/400 (1%)
Frame = +1
Query: 7 SIDILKNQDVIRSVLNILQTNTSVASSLGPHFFPQISLIFLDMLTVYRMYSELVSSTIAE 186
++DILK+ + ++ + +IL+TN ++G F Q+ I+LDML VY+ SE +S+ I
Sbjct: 675 NVDILKDPETVKQLGSILKTNVRACKAVGHPFVIQLGRIYLDMLNVYKCLSENISAAIQA 734
Query: 187 GGPFASRTSFVKLLRSVKRETLKLIETFVDKAEDLPHVGKQFVPPMMDPVLGDYARNVPD 366
G ++ ++ +R+VKRETLKLI +V ++ D V + FVPP++D VL DY RNVP
Sbjct: 735 NGEMVTKQPLIRSMRTVKRETLKLISGWVSRSNDPQMVAENFVPPLLDAVLIDYQRNVPA 794
Query: 367 ARESEVLSLFATIINKYKGEMLEDVPRIFEAVFQCTLEMITKNFEDYPEHRLKFFSLLRA 546
ARE EVLS A I+NK G + ++P+IF+AVF+CTL MI K+FE+YPEHR FF LL+A
Sbjct: 795 AREPEVLSTMAIIVNKLGGHITAEIPQIFDAVFECTLNMINKDFEEYPEHRTNFFLLLQA 854
Query: 547 IGTHCFQALIQLSSQQLKLVIDSINWAFRHTERNIAETGLSLLLEILKKF-QASGFQNQF 723
+ +HCF A + + Q KLV+DSI WAF+HT RN+A+TGL +L +L+ Q F
Sbjct: 855 VNSHCFPAFLAIPPTQFKLVLDSIIWAFKHTMRNVADTGLQILFTLLQNVAQEEAAAQSF 914
Query: 724 YKTYFLTIEQEIFAVLTDTFHKPGFKLHVSVLQHLFCAVDGLTEPLWDASSVPYQYTDNA 903
Y+TYF I Q IF+V+TDT H G +H S+L ++F V+ E S P +N
Sbjct: 915 YQTYFCDILQHIFSVVTDTSHTAGLTMHASILAYMFNLVE---EGKISTSLNPGNPVNNQ 971
Query: 904 MFVRDYTIKLLGTSFPNMTVAEVTKFVDGLLSSKLDLPSFKNHIRDFLVQSKEFSAQDNK 1083
+F+++Y LL ++FP++ A+V FV GL S D+P+FK H+RDFLVQ KEF+ +D
Sbjct: 972 IFLQEYVANLLKSAFPHLQDAQVKLFVTGLFSLNQDIPAFKEHLRDFLVQIKEFAGEDTS 1031
Query: 1084 DLY----XXXXXXXXXXXXXXMLAIPGLIAPSELQDEMVD 1191
DL+ +++PG+ P E+ +EM D
Sbjct: 1032 DLFLEEREIALRQADEEKHKRQMSVPGIFNPHEIPEEMCD 1071
>sptr|Q9PW90|Q9PW90 CRM1/XPO1 protein.
Length = 1071
Score = 329 bits (844), Expect = 7e-89
Identities = 169/402 (42%), Positives = 252/402 (62%), Gaps = 7/402 (1%)
Frame = +1
Query: 7 SIDILKNQDVIRSVLNILQTNTSVASSLGPHFFPQISLIFLDMLTVYRMYSELVSSTIAE 186
++DILK+ + ++ + +IL+TN ++G F Q+ I+LDML VY+ SE +S+ I
Sbjct: 675 NVDILKDPETVKQLGSILKTNVRACKAVGHPFVIQLGRIYLDMLNVYKCLSENISAAIQA 734
Query: 187 GGPFASRTSFVKLLRSVKRETLKLIETFVDKAEDLPHVGKQFVPPMMDPVLGDYARNVPD 366
G ++ ++ +R+VKRETLKLI +V ++ D V + FVPP++D VL DY RNVP
Sbjct: 735 NGEMVTKQPLIRSMRTVKRETLKLISGWVSRSSDPQMVAENFVPPLLDAVLIDYQRNVPA 794
Query: 367 ARESEVLSLFATIINKYKGEMLEDVPRIFEAVFQCTLEMITKNFEDYPEHRLKFFSLLRA 546
ARE EVLS ATI+NK + ++P+IF+AVF+CTL MI K+FE+YPEHR FF LL+A
Sbjct: 795 AREPEVLSTMATIVNKLGVHITAEIPQIFDAVFECTLNMINKDFEEYPEHRTHFFLLLQA 854
Query: 547 IGTHCFQALIQLSSQQLKLVIDSINWAFRHTERNIAETGLSLLLEILKKF-QASGFQNQF 723
+ +HCF A + + Q KLV+DSI WAF+HT RN+A+TGL +L +L+ Q F
Sbjct: 855 VNSHCFPAFLAIPPAQFKLVLDSIIWAFKHTMRNVADTGLQILYTLLQNVAQEEAAAQSF 914
Query: 724 YKTYFLTIEQEIFAVLTDTFHKPGFKLHVSVLQHLFCAVD--GLTEPLWDASSVPYQYTD 897
Y+TYF I Q F+V+TDT H G +H S+L ++F V+ + PL AS + +
Sbjct: 915 YQTYFCDILQHTFSVVTDTSHTAGLTMHASILAYMFNLVEEGKINTPLNQASPL-----N 969
Query: 898 NAMFVRDYTIKLLGTSFPNMTVAEVTKFVDGLLSSKLDLPSFKNHIRDFLVQSKEFSAQD 1077
N +F+++Y LL ++FP++ A+V FV GL S D+ +FK H+RDFLVQ KE++ +D
Sbjct: 970 NQLFIQEYVANLLKSAFPHLQDAQVKLFVTGLFSLNQDIAAFKEHLRDFLVQIKEYAGED 1029
Query: 1078 NKDLY----XXXXXXXXXXXXXXMLAIPGLIAPSELQDEMVD 1191
DL+ +++PG++ P E+ +EM D
Sbjct: 1030 TSDLFLEERESSLRQAQEEKHKLQMSVPGILNPHEIPEEMCD 1071
>sw|P14068|XPO1_SCHPO Exportin 1 (Chromosome region maintenance
protein 1) (Caffeine resistance protein 2).
Length = 1078
Score = 313 bits (803), Expect = 4e-84
Identities = 161/394 (40%), Positives = 251/394 (63%), Gaps = 8/394 (2%)
Frame = +1
Query: 16 ILKNQDVIRSVLNILQTNTSVASSLGPHFFPQISLIFLDMLTVYRMYSELVSSTIAEGGP 195
+L + ++ + N+L+TN + +S+G F+PQI+ ++DML +Y+ S L+S +A G
Sbjct: 679 VLGDPQTVKILANVLKTNVAACTSIGSGFYPQIAKNYVDMLGLYKAVSGLISEVVAAQGN 738
Query: 196 FASRTSFVKLLRSVKRETLKLIETFVDKAEDLPHVGKQFVPPMMDPVLGDYARNVPDARE 375
A++T V+ LR++K+E LKL++ ++ +AEDL VG +P + + VL DY +NVPDAR+
Sbjct: 739 IATKTPHVRGLRTIKKEILKLVDAYISRAEDLELVGNTLIPALFEAVLLDYLQNVPDARD 798
Query: 376 SEVLSLFATIINKYKGEMLEDVPRIFEAVFQCTLEMITKNFEDYPEHRLKFFSLLRAIGT 555
+EVL+L TI+N+ + + +P + +AVF CTLEMI+K+F +YPEHR FF LLRAI
Sbjct: 799 AEVLNLITTIVNQLSELLTDKIPLVLDAVFGCTLEMISKDFSEYPEHRAAFFQLLRAINL 858
Query: 556 HCFQALIQLSSQQLKLVIDSINWAFRHTERNIAETGLSLLLEILKKFQASG--FQNQFYK 729
+CF AL+ + + Q KLVI+SI W+F+H R+I ETGL++LLE++ + G N F++
Sbjct: 859 NCFPALLNIPAPQFKLVINSIVWSFKHVSRDIQETGLNILLELINNMASMGPDVSNAFFQ 918
Query: 730 TYFLTIEQEIFAVLTDTFHKPGFKLHVSVLQHLFCAVDG--LTEPLWDASSVPYQYTDNA 903
TY++++ Q+I VLTD+ HK GFKL +L LF V+ +T PL+D S P Q +N
Sbjct: 919 TYYISLLQDILYVLTDSDHKSGFKLQSLILARLFYLVESNQITVPLYDPSQFP-QEMNNQ 977
Query: 904 MFVRDYTIKLLGTSFPNMTVAEVTKFVDGLLSSKLDLPSFKNHIRDFLVQSKEFSAQDNK 1083
+F+R Y + LL T+FP++ ++ +FV +L+ D FK +RDFL+Q KEF DN
Sbjct: 978 LFLRQYIMNLLVTAFPHLQPIQIQEFVQTVLALNQDSIKFKLALRDFLIQLKEFGG-DNA 1036
Query: 1084 DLYXXXXXXXXXXXXXXML----AIPGLIAPSEL 1173
+LY L +PG+I P ++
Sbjct: 1037 ELYLEEKEQELAAQQKAQLEKAMTVPGMIKPVDM 1070
>sptr|Q7RWC2|Q7RWC2 Hypothetical protein.
Length = 1083
Score = 307 bits (787), Expect = 3e-82
Identities = 171/407 (42%), Positives = 245/407 (60%), Gaps = 13/407 (3%)
Frame = +1
Query: 1 SLSIDILKNQDVIRSVLNILQTNTSVASSLGPHFFPQISLIFLDMLTVYRMYSELVSSTI 180
SL+ +IL++ D I+ + NI++TN S SS+G +FFPQI ++ DML +Y S+L+S +
Sbjct: 676 SLNPNILQDADTIKVIGNIMKTNVSACSSIGTYFFPQIGNLYSDMLQMYAATSQLISEAV 735
Query: 181 AEGGPFASRTSFVKLLRSVKRETLKLIETFVDKAEDLPHVGKQFVPPMMDPVLGDYARNV 360
A G A++ V+ LR++K+E LKLIET+VDKAEDL V +Q VPP++D VL DY RNV
Sbjct: 736 AREGEIATKMPKVRGLRTIKKEILKLIETYVDKAEDLQAVREQMVPPLLDSVLVDYNRNV 795
Query: 361 PDARESEVLSLFATIINKYKGEMLEDVPRIFEAVFQCTLEMITKNFEDYPEHRLKFFSLL 540
P AR++EVL +I K M + VP I E VF+CTL+MI KNF +YPEHR++FF+LL
Sbjct: 796 PGARDAEVLRAMTAMITKLSALMEDQVPIIMENVFECTLDMINKNFSEYPEHRVEFFNLL 855
Query: 541 RAIGTHCFQALIQLSSQQLKLVIDSINWAFRHTERNIAETGLSLLLEILKKFQAS---GF 711
RAI HCF AL++L ++Q K VIDS WA +H R++ GL++ LE++
Sbjct: 856 RAINLHCFPALLKLDNRQFKFVIDSCMWASKHDNRDVETAGLNMCLELVNNIAEKTDVQT 915
Query: 712 QNQFYKTYFLTIEQEIFAVLTDTFHKPGFKLHVSVLQHLFCAV---DGLTEPLWDASSVP 882
N F+ +F++I Q++F VLTD HK GFK +L +F V DG + P
Sbjct: 916 SNAFFNQFFVSILQDVFFVLTDQDHKAGFKTQSMLLMRMFYFVHPADGSPSRIQGPIYQP 975
Query: 883 YQY---TDNAMFVRDYTIKLLGTSFPNMTVAEVTKFVDGLLSSKLDLPSFKNHIRDFLVQ 1053
Q T N F+ + LL T+F N+T A++T FV+GL + F+ +RDFL+
Sbjct: 976 DQAQPGTSNKEFLTMFVGNLLQTAFANLTPAQITSFVEGLFTLNTQYDKFRLALRDFLIS 1035
Query: 1054 SKEFSAQDNKDLYXXXXXXXXXXXXXXML----AIPGLIAPSELQDE 1182
+EF A DN +LY + + GL+ PSEL+D+
Sbjct: 1036 LREF-AGDNAELYLLEKEQQETAAKAADIERRSKVSGLLKPSELEDD 1081
>sptrnew|CAE55861|CAE55861 Exportin 1.
Length = 1054
Score = 301 bits (772), Expect = 1e-80
Identities = 157/401 (39%), Positives = 242/401 (60%), Gaps = 8/401 (1%)
Frame = +1
Query: 13 DILKNQDVIRSVLNILQTNTSVASSLGPHFFPQISLIFLDMLTVYRMYSELVSSTIAEGG 192
DILK+ ++ + +IL+TN +LG ++ Q+ I+LDML VY++ SE +S IA G
Sbjct: 660 DILKDMAAVKQIGSILKTNVRACKALGHNYVIQLGCIYLDMLNVYKIMSENISQAIAING 719
Query: 193 PFASRTSFVKLLRSVKRETLKLIETFVDKAEDLPHVGKQFVPPMMDPVLGDYARN-VPDA 369
+ +K + VK+ETL LI ++ ++ D V + F+PP++D VL DY R VP A
Sbjct: 720 ITINNQPLIKAMHVVKKETLTLISEWIVRSNDAQMVMESFIPPLLDAVLMDYQRTIVPAA 779
Query: 370 RESEVLSLFATIINKYKGEMLEDVPRIFEAVFQCTLEMITKNFEDYPEHRLKFFSLLRAI 549
RE VLS A I+NK +G + +VP+IF+AVF+CTL+MI KNFEDYP+HR F+ LL+A+
Sbjct: 780 REPTVLSTMAAIVNKLEGVITSEVPKIFDAVFECTLDMINKNFEDYPQHRTYFYELLQAV 839
Query: 550 GTHCFQALIQLSSQQLKLVIDSINWAFRHTERNIAETGLSLLLEILKKF-QASGFQNQFY 726
HCF+A + + Q KLV DSI W F+HT RN+A+TGLS+L ++L+ Q FY
Sbjct: 840 NMHCFKAFLNIPPAQFKLVFDSIVWGFKHTMRNVADTGLSILFQLLQNLEQHPQAAQSFY 899
Query: 727 KTYFLTIEQEIFAVLTDTFHKPGFKLHVSVLQHLFCAVD--GLTEPLWDASSVPYQYTDN 900
+TYF I +IF+V+TDT H + H +L H+F V+ +T L +P +N
Sbjct: 900 QTYFTDILMQIFSVVTDTSHTASLQSHAQILAHMFSLVEANAITVAL---GPIP----NN 952
Query: 901 AMFVRDYTIKLLGTSFPNMTVAEVTKFVDGLLSSKLDLPSFKNHIRDFLVQSKEFSAQDN 1080
+++++Y LL ++F ++T ++ FV GL + D+ +FK H+RDF++Q +E + D+
Sbjct: 953 VLYIQEYVASLLKSAFSHLTDNQIKVFVTGLFNLDHDVAAFKEHLRDFIIQIREVTGDDD 1012
Query: 1081 KDLY----XXXXXXXXXXXXXXMLAIPGLIAPSELQDEMVD 1191
DLY + +PGL+ P E+ ++M D
Sbjct: 1013 SDLYLDEREQELKQAQDEKRRHQMTVPGLLNPHEIPEDMQD 1053
>sptr|Q8IH79|Q8IH79 GH01059p (Fragment).
Length = 697
Score = 295 bits (756), Expect = 1e-78
Identities = 154/403 (38%), Positives = 242/403 (60%), Gaps = 6/403 (1%)
Frame = +1
Query: 1 SLSIDILKNQDVIRSVLNILQTNTSVASSLGPHFFPQISLIFLDMLTVYRMYSELVSSTI 180
S ++D LKN ++ + +IL+TN + +LG + Q+ I+LDML VY++ SE + I
Sbjct: 299 SKNVDFLKNMTAVKQLGSILKTNVAACKALGHAYVIQLGRIYLDMLNVYKITSENIIQAI 358
Query: 181 AEGGPFASRTSFVKLLRSVKRETLKLIETFVDKAEDLPHVGKQFVPPMMDPVLGDYAR-N 357
G + +K + VK+ETL LI +V ++ D V F+PP++D +L DY R
Sbjct: 359 EVNGVNVNNQPLIKTMHVVKKETLNLISEWVSRSNDNQLVMDNFIPPLLDAILLDYQRCK 418
Query: 358 VPDARESEVLSLFATIINKYKGEMLEDVPRIFEAVFQCTLEMITKNFEDYPEHRLKFFSL 537
VP ARE +VLS A I++K + + +VP+IF+AVF+CTL+MI KNFED+P+HRL F+ L
Sbjct: 419 VPSAREPKVLSAMAIIVHKLRQHITNEVPKIFDAVFECTLDMINKNFEDFPQHRLSFYEL 478
Query: 538 LRAIGTHCFQALIQLSSQQLKLVIDSINWAFRHTERNIAETGLSLLLEILKKF-QASGFQ 714
L+A+ HCF+A + + Q KLV DS+ WAF+HT RN+A+ GL++L ++L+ Q G
Sbjct: 479 LQAVNAHCFKAFLNIPPAQFKLVFDSVVWAFKHTMRNVADMGLNILFKMLQNLDQHPGAA 538
Query: 715 NQFYKTYFLTIEQEIFAVLTDTFHKPGFKLHVSVLQHLFCAVDGLTEPLWDASSVPYQYT 894
FY+TYF I +IF+V+TDT H G H +L ++F V+ + + +P
Sbjct: 539 QSFYQTYFTDILMQIFSVVTDTSHTAGLPNHAIILAYMFSLVENRKITV-NLGPIP---- 593
Query: 895 DNAMFVRDYTIKLLGTSFPNMTVAEVTKFVDGLLSSKLDLPSFKNHIRDFLVQSKEFSAQ 1074
DN +F+++Y LL ++F +++ +V FV GL + ++ +FK H+RDFL+Q +E + +
Sbjct: 594 DNMIFIQEYVASLLKSAFTHLSDNQVKVFVTGLFNLDENVQAFKEHLRDFLIQIREATGE 653
Query: 1075 DNKDLY----XXXXXXXXXXXXXXMLAIPGLIAPSELQDEMVD 1191
D+ DLY IPG++ P EL ++M D
Sbjct: 654 DDSDLYLEEREAALAEEQSNKHQMQRNIPGMLNPHELPEDMQD 696
>sptr|Q9TVM2|Q9TVM2 Chromosomal region maintenance 1 protein
(LD45706p) (CG13387-PA).
Length = 1063
Score = 295 bits (756), Expect = 1e-78
Identities = 154/403 (38%), Positives = 242/403 (60%), Gaps = 6/403 (1%)
Frame = +1
Query: 1 SLSIDILKNQDVIRSVLNILQTNTSVASSLGPHFFPQISLIFLDMLTVYRMYSELVSSTI 180
S ++D LKN ++ + +IL+TN + +LG + Q+ I+LDML VY++ SE + I
Sbjct: 665 SKNVDFLKNMTAVKQLGSILKTNVAACKALGHAYVIQLGRIYLDMLNVYKITSENIIQAI 724
Query: 181 AEGGPFASRTSFVKLLRSVKRETLKLIETFVDKAEDLPHVGKQFVPPMMDPVLGDYAR-N 357
G + +K + VK+ETL LI +V ++ D V F+PP++D +L DY R
Sbjct: 725 EVNGVNVNNQPLIKTMHVVKKETLNLISEWVSRSNDNQLVMDNFIPPLLDAILLDYQRCK 784
Query: 358 VPDARESEVLSLFATIINKYKGEMLEDVPRIFEAVFQCTLEMITKNFEDYPEHRLKFFSL 537
VP ARE +VLS A I++K + + +VP+IF+AVF+CTL+MI KNFED+P+HRL F+ L
Sbjct: 785 VPSAREPKVLSAMAIIVHKLRQHITNEVPKIFDAVFECTLDMINKNFEDFPQHRLSFYEL 844
Query: 538 LRAIGTHCFQALIQLSSQQLKLVIDSINWAFRHTERNIAETGLSLLLEILKKF-QASGFQ 714
L+A+ HCF+A + + Q KLV DS+ WAF+HT RN+A+ GL++L ++L+ Q G
Sbjct: 845 LQAVNAHCFKAFLNIPPAQFKLVFDSVVWAFKHTMRNVADMGLNILFKMLQNLDQHPGAA 904
Query: 715 NQFYKTYFLTIEQEIFAVLTDTFHKPGFKLHVSVLQHLFCAVDGLTEPLWDASSVPYQYT 894
FY+TYF I +IF+V+TDT H G H +L ++F V+ + + +P
Sbjct: 905 QSFYQTYFTDILMQIFSVVTDTSHTAGLPNHAIILAYMFSLVENRKITV-NLGPIP---- 959
Query: 895 DNAMFVRDYTIKLLGTSFPNMTVAEVTKFVDGLLSSKLDLPSFKNHIRDFLVQSKEFSAQ 1074
DN +F+++Y LL ++F +++ +V FV GL + ++ +FK H+RDFL+Q +E + +
Sbjct: 960 DNMIFIQEYVASLLKSAFTHLSDNQVKVFVTGLFNLDENVQAFKEHLRDFLIQIREATGE 1019
Query: 1075 DNKDLY----XXXXXXXXXXXXXXMLAIPGLIAPSELQDEMVD 1191
D+ DLY IPG++ P EL ++M D
Sbjct: 1020 DDSDLYLEEREAALAEEQSNKHQMQRNIPGMLNPHELPEDMQD 1062
>sptr|Q9U699|Q9U699 Chromosomal region maintenance protein CRM1.
Length = 1063
Score = 295 bits (756), Expect = 1e-78
Identities = 154/403 (38%), Positives = 242/403 (60%), Gaps = 6/403 (1%)
Frame = +1
Query: 1 SLSIDILKNQDVIRSVLNILQTNTSVASSLGPHFFPQISLIFLDMLTVYRMYSELVSSTI 180
S ++D LKN ++ + +IL+TN + +LG + Q+ I+LDML VY++ SE + I
Sbjct: 665 SKNVDFLKNMTAVKQLGSILKTNVAACKALGHAYVIQLGRIYLDMLNVYKITSENIIQAI 724
Query: 181 AEGGPFASRTSFVKLLRSVKRETLKLIETFVDKAEDLPHVGKQFVPPMMDPVLGDYAR-N 357
G + +K + VK+ETL LI +V ++ D V F+PP++D +L DY R
Sbjct: 725 EVNGVNVNNQPLIKTMHVVKKETLNLISEWVSRSNDNQLVMDNFIPPLLDAILLDYQRCK 784
Query: 358 VPDARESEVLSLFATIINKYKGEMLEDVPRIFEAVFQCTLEMITKNFEDYPEHRLKFFSL 537
VP ARE +VLS A I++K + + +VP+IF+AVF+CTL+MI KNFED+P+HRL F+ L
Sbjct: 785 VPSAREPKVLSAMAIIVHKLRQHITNEVPKIFDAVFECTLDMINKNFEDFPQHRLSFYEL 844
Query: 538 LRAIGTHCFQALIQLSSQQLKLVIDSINWAFRHTERNIAETGLSLLLEILKKF-QASGFQ 714
L+A+ HCF+A + + Q KLV DS+ WAF+HT RN+A+ GL++L ++L+ Q G
Sbjct: 845 LQAVNAHCFKAFLNIPPAQFKLVFDSVVWAFKHTMRNVADMGLNILFKMLQNLDQHPGAA 904
Query: 715 NQFYKTYFLTIEQEIFAVLTDTFHKPGFKLHVSVLQHLFCAVDGLTEPLWDASSVPYQYT 894
FY+TYF I +IF+V+TDT H G H +L ++F V+ + + +P
Sbjct: 905 QSFYQTYFTDILMQIFSVVTDTSHTAGLPNHAIILAYMFSLVENRKITV-NLGPIP---- 959
Query: 895 DNAMFVRDYTIKLLGTSFPNMTVAEVTKFVDGLLSSKLDLPSFKNHIRDFLVQSKEFSAQ 1074
DN +F+++Y LL ++F +++ +V FV GL + ++ +FK H+RDFL+Q +E + +
Sbjct: 960 DNMIFIQEYVASLLKSAFTHLSDNQVKVFVTGLFNLDENVQAFKEHLRDFLIQIREAAGE 1019
Query: 1075 DNKDLY----XXXXXXXXXXXXXXMLAIPGLIAPSELQDEMVD 1191
D+ DLY IPG++ P EL ++M D
Sbjct: 1020 DDSDLYLEEREAALAEEQSNKHQMQRNIPGMLNPHELPEDMQD 1062
>sptr|Q7PMB7|Q7PMB7 ENSANGP00000013197 (Fragment).
Length = 1057
Score = 291 bits (745), Expect = 2e-77
Identities = 150/403 (37%), Positives = 247/403 (61%), Gaps = 8/403 (1%)
Frame = +1
Query: 7 SIDILKNQDVIRSVLNILQTNTSVASSLGPHFFPQISLIFLDMLTVYRMYSELVSSTIAE 186
++DILK+ ++ + +IL+TN +LG + Q+ I+LDML VY++ SE ++ I+
Sbjct: 661 NVDILKDMGAVKQLGSILKTNVRACKALGHSYVSQLGRIYLDMLNVYKIMSENITQAISL 720
Query: 187 GGPFASRTSFVKLLRSVKRETLKLIETFVDKAEDLPHVGKQFVPPMMDPVLGDYARN-VP 363
G + +K + VK+ETL LI +V K+ D V + F+PP+++ VL DY R VP
Sbjct: 721 NGLSINNQPLIKAMHVVKKETLTLISEWVWKSNDPKMVMENFIPPLLEAVLFDYQRTKVP 780
Query: 364 DARESEVLSLFATIINKYKGEMLEDVPRIFEAVFQCTLEMITKNFEDYPEHRLKFFSLLR 543
+ARE VLS A+I+N+ + + ++P+IF+A+F CTL+MI KNFEDYP+HR F+ LL+
Sbjct: 781 NAREPLVLSTMASIVNRLQAVITPEIPKIFDALFDCTLDMINKNFEDYPQHRTNFYELLQ 840
Query: 544 AIGTHCFQALIQLSSQQLKLVIDSINWAFRHTERNIAETGLSLLLEILKKF-QASGFQNQ 720
A+ +CF+A + + +Q KLV DSI WAF+HT RN+A+TGL++L+++L+ Q
Sbjct: 841 AVNMYCFKAFLSIPPEQFKLVFDSIVWAFKHTMRNVADTGLNILMQMLQNLEQHPQAAQS 900
Query: 721 FYKTYFLTIEQEIFAVLTDTFHKPGFKLHVSVLQHLFCAVDG--LTEPLWDASSVPYQYT 894
FY+TY+ I +IF+V+TDT H + H ++L ++F V+ +T L +
Sbjct: 901 FYQTYYTDILMQIFSVVTDTSHTASLQNHATILAYMFSLVEAGRITVKLGPSD------- 953
Query: 895 DNAMFVRDYTIKLLGTSFPNMTVAEVTKFVDGLLSSKLDLPSFKNHIRDFLVQSKEFSAQ 1074
DN + +++Y LL ++F ++T ++ FV GL + D+ +FK H+RDFL+Q KE + +
Sbjct: 954 DNVLNIQEYVAMLLKSAFSHLTGNQIKIFVTGLFNLDQDVHAFKEHLRDFLIQIKEVTGE 1013
Query: 1075 DNKDLY----XXXXXXXXXXXXXXMLAIPGLIAPSELQDEMVD 1191
D+ DLY ++ +PG++ P E+ ++M D
Sbjct: 1014 DDSDLYLEERENELKKIQEEKRRMLMTVPGMMNPHEMPEDMQD 1056
>sptr|Q872T2|Q872T2 Probable nuclear export factor CRM1 (Fragment).
Length = 759
Score = 288 bits (738), Expect = 1e-76
Identities = 165/404 (40%), Positives = 234/404 (57%), Gaps = 10/404 (2%)
Frame = +1
Query: 1 SLSIDILKNQDVIRSVLNILQTNTSVASSLGPHFFPQISLIFLDMLTVYRMYSELVSSTI 180
SL+ +IL++ D I+ + NI++TN S SS+G +FFPQI ++ DML +Y S+L+S +
Sbjct: 368 SLNPNILQDADTIKVIGNIMKTNVSACSSIGTYFFPQIGNLYSDMLQMYAATSQLISEAV 427
Query: 181 AEGGPFASRTSFVKLLRSVKRETLKLIETFVDKAEDLPHVGKQFVPPMMDPVLGDYARNV 360
A G A++ V+ LR++K+E LKLIET+VDKAEDL V +Q VPP++D VL DY RNV
Sbjct: 428 AREGEIATKMPKVRGLRTIKKEILKLIETYVDKAEDLQAVREQMVPPLLDSVLVDYNRNV 487
Query: 361 PDARESEVLSLFATIINKYKGEMLEDVPRIFEAVFQCTLEMITKNFEDYPEHRLKFFSLL 540
P AR++EVL +I K M + VP I E VF+CTL+MI KNF +YPEHR++FF+LL
Sbjct: 488 PGARDAEVLRAMTAMITKLSALMEDQVPIIMENVFECTLDMINKNFSEYPEHRVEFFNLL 547
Query: 541 RAIGTHCFQALIQLSSQQLKLVIDSINWAFRHTERNIAETGLSLLLEILKKFQAS---GF 711
RAI HCF AL++L ++Q K VIDS WA +H R++ GL++ LE++
Sbjct: 548 RAINLHCFPALLKLDNRQFKFVIDSCMWASKHDNRDVETAGLNMCLELVNNIAEKTDVQT 607
Query: 712 QNQFYKTYFLTIEQEIFAVLTDTFHKPGFKLHVSVLQHLFCAV---DGLTEPLWDASSVP 882
N F+ +F++I Q++F VLTD HK GFK +L +F V DG + P
Sbjct: 608 SNAFFNQFFVSILQDVFFVLTDQDHKAGFKTQSMLLMRMFYFVHPADGSPSRI----QGP 663
Query: 883 YQYTDNAMFVRDYTIKLLGTSFPNMTVAEVTKFVDGLLSSKLDLPSFKNHIRDFLVQSKE 1062
D A GTS +T FV+GL + F+ +RDFL+ +E
Sbjct: 664 IYQPDQAQ---------PGTSNKEFLTMFITSFVEGLFTLNTQYDKFRLALRDFLISLRE 714
Query: 1063 FSAQDNKDLYXXXXXXXXXXXXXXML----AIPGLIAPSELQDE 1182
F A DN +LY + + GL+ PSEL+D+
Sbjct: 715 F-AGDNAELYLLEKEQQETAAKAADIERRSKVSGLLKPSELEDD 757
>sw|P30822|XPO1_YEAST Exportin 1 (Chromosome region maintenance
protein 1).
Length = 1084
Score = 274 bits (700), Expect = 3e-72
Identities = 145/397 (36%), Positives = 229/397 (57%), Gaps = 9/397 (2%)
Frame = +1
Query: 16 ILKNQDVIRSVLNILQTNTSVASSLGPHFFPQISLIFLDMLTVYRMYSELVSSTIAEGGP 195
+L + + ++ + NI++TN +V +S+G F+PQ+ I+ +ML +YR S ++S+ +A G
Sbjct: 689 LLLDSETVKIIANIIKTNVAVCTSMGADFYPQLGHIYYNMLQLYRAVSSMISAQVAAEGL 748
Query: 196 FASRTSFVKLLRSVKRETLKLIETFVDKAEDLPHVGKQFVPPMMDPVLGDYARNVPDARE 375
A++T V+ LR++K+E LKL+ET++ KA +L V K V P+++ VL DY NVPDAR+
Sbjct: 749 IATKTPKVRGLRTIKKEILKLVETYISKARNLDDVVKVLVEPLLNAVLEDYMNNVPDARD 808
Query: 376 SEVLSLFATIINKYKGEMLEDVPRIFEAVFQCTLEMITKNFEDYPEHRLKFFSLLRAIGT 555
+EVL+ T++ K + + V I ++VF+CTL+MI K+F +YPEHR++F+ LL+ I
Sbjct: 809 AEVLNCMTTVVEKVGHMIPQGVILILQSVFECTLDMINKDFTEYPEHRVEFYKLLKVINE 868
Query: 556 HCFQALIQLSSQQLKLVIDSINWAFRHTERNIAETGLSLLLEILKKFQASG---FQNQFY 726
F A ++L KL +D+I WAF+H R++ GL + L+++K + G F N+F+
Sbjct: 869 KSFAAFLELPPAAFKLFVDAICWAFKHNNRDVEVNGLQIALDLVKNIERMGNVPFANEFH 928
Query: 727 KTYFLTIEQEIFAVLTDTFHKPGFKLHVSVLQHLFCAV--DGLTEPLWDASSVPYQYTDN 900
K YF E F VLTD+ HK GF +L L V + ++ PL+ + VP Q T N
Sbjct: 929 KNYFFIFVSETFFVLTDSDHKSGFSKQALLLMKLISLVYDNKISVPLYQEAEVP-QGTSN 987
Query: 901 AMFVRDYTIKLLGTSFPNMTVAEVTKFVDGLLSSKLDLPSFKNHIRDFLVQSKEFSAQDN 1080
+++ Y +L +FP++T ++ F+ L DL FK +RDFLVQ KE
Sbjct: 988 QVYLSQYLANMLSNAFPHLTSEQIASFLSALTKQYKDLVVFKGTLRDFLVQIKEVGGDPT 1047
Query: 1081 KDLYX----XXXXXXXXXXXXXMLAIPGLIAPSELQD 1179
L+ I GL+ PSEL D
Sbjct: 1048 DYLFAEDKENALMEQNRLEREKAAKIGGLLKPSELDD 1084
>sptr|Q9P8X1|Q9P8X1 Crm1p.
Length = 1079
Score = 260 bits (664), Expect = 5e-68
Identities = 136/399 (34%), Positives = 233/399 (58%), Gaps = 9/399 (2%)
Frame = +1
Query: 13 DILKNQDVIRSVLNILQTNTSVASSLGPHFFPQISLIFLDMLTVYRMYSELVSSTIAEGG 192
++L N + ++ + NI++TN +V +LGP F+ Q+ +++DML++Y+ S+++S +A+ G
Sbjct: 682 ELLSNTETVKIIANIIKTNVAVCKALGPGFYSQLGGLYVDMLSLYKAVSQMISDAVAKDG 741
Query: 193 PFASRTSFVKLLRSVKRETLKLIETFVDKAEDLPHVGKQFVPPMMDPVLGDYARNVPDAR 372
A++T V+ LR++K+E LK+IET++++A++L + + V P+ VL DY+ NVPDAR
Sbjct: 742 IIATKTPKVRGLRTIKKEILKMIETYINQADNLQEIVRDLVQPLFGAVLEDYSSNVPDAR 801
Query: 373 ESEVLSLFATIINKYKGEMLEDVPRIFEAVFQCTLEMITKNFEDYPEHRLKFFSLLRAIG 552
++EVL +++K + + V I + VF+CTL+MI +F +YPEHR++F+ LL+ I
Sbjct: 802 DAEVLRCLTALVSKAGHLIPDGVVLILQNVFECTLDMIKNDFVEYPEHRVEFYKLLKEIN 861
Query: 553 THCFQALIQLSSQQLKLVIDSINWAFRHTERNIAETGLSLLLEILKKFQASG---FQNQF 723
FQ L+QLS + + +I++ WAF+H R + + GLSL LE+++ + G F F
Sbjct: 862 AKSFQGLLQLSGEAFQSLINAALWAFKHNNREVEDNGLSLTLELIENVEKLGDTPFTKAF 921
Query: 724 YKTYFLTIEQEIFAVLTDTFHKPGFKLHVSVLQHLFCAVDG--LTEPLWDASSVPYQYTD 897
Y+ ++ I + V T HK GF+ +L L V+ + PL+ + P + T
Sbjct: 922 YENFYFQILSDTLYVFTQPDHKAGFRYQAQLLAQLIHLVEDNVIKYPLYTSDQAP-EGTS 980
Query: 898 NAMFVRDYTIKLLGTSFPNMTVAEVTKFVDGLLSSKLDLPSFKNHIRDFLVQSKEFSAQD 1077
N+ F++ Y +LL ++F N+ ++ F+ L + DL FK+ +RDFLVQ KEF
Sbjct: 981 NSDFLKQYLSQLLSSAFDNLQEVQLINFLKVLTTVYNDLFKFKSVLRDFLVQLKEFGGDP 1040
Query: 1078 NKDLYXXXXXXXXXXXXXXM----LAIPGLIAPSELQDE 1182
L+ + + GLI PSE+ DE
Sbjct: 1041 TDYLFAEDKQIEREEQDRLQRERDMQVGGLIRPSEMDDE 1079
>sptr|Q23089|Q23089 Hypothetical protein.
Length = 1080
Score = 255 bits (652), Expect = 1e-66
Identities = 141/399 (35%), Positives = 221/399 (55%), Gaps = 5/399 (1%)
Frame = +1
Query: 1 SLSIDILKNQDVIRSVLNILQTNTSVASSLGPHFFPQISLIFLDMLTVYRMYSELVSSTI 180
S + +L+ ++++SVLNIL+TN + S+G F Q+ I+ D+L++Y++ SE VS +
Sbjct: 683 STNDSVLEEPEMVKSVLNILKTNVAACKSIGSSFVTQLGNIYSDLLSLYKILSEKVSRAV 742
Query: 181 AEGGPFASRTSFVKLLRSVKRETLKLIETFVDKAEDLPHVGKQFVPPMMDPVLGDYARNV 360
G A + VK +R+VKRE L L+ TF+ K D + VPP+ D VL DY +NV
Sbjct: 743 TTAGEEALKNPLVKTMRAVKREILILLSTFISKNGDAKLILDSIVPPLFDAVLFDYQKNV 802
Query: 361 PDARESEVLSLFATIINKYKGEMLEDVPRIFEAVFQCTLEMITKNFEDYPEHRLKFFSLL 540
P ARE +VLSL + ++ + + VP I AVFQC+++MI K+ E +PEHR FF L+
Sbjct: 803 PQAREPKVLSLLSILVTQLGSLLCPQVPSILSAVFQCSIDMINKDMEAFPEHRTNFFELV 862
Query: 541 RAIGTHCFQALIQLSSQQLKLVIDSINWAFRHTERNIAETGLSLLLEILKKF--QASGFQ 714
++ CF +++ + L VID++ WAF+HT RN+AE GL +L E+L + Q
Sbjct: 863 LSLVQECFPVFMEMPPEDLGTVIDAVVWAFQHTMRNVAEIGLDILKELLARVSEQDDKIA 922
Query: 715 NQFYKTYFLTIEQEIFAVLTDT--FHKPGFKLHVSVLQHLFCAVD-GLTEPLWDASSVPY 885
FYK Y++ + + + AV D+ H G + VL LF A + + PL DA+ P
Sbjct: 923 QPFYKRYYIDLLKHVLAVACDSSQVHVAGLTYYAEVLCALFRAPEFSIKVPLNDAN--PS 980
Query: 886 QYTDNAMFVRDYTIKLLGTSFPNMTVAEVTKFVDGLLSSKLDLPSFKNHIRDFLVQSKEF 1065
Q N ++ ++ F NM ++ + G S ++ S +NH+RDFL+Q KE
Sbjct: 981 Q--PNIDYIYEHIGGNFQAHFDNMNQDQIRIIIKGFFSFNTEISSMRNHLRDFLIQIKEH 1038
Query: 1066 SAQDNKDLYXXXXXXXXXXXXXXMLAIPGLIAPSELQDE 1182
+ +D DLY +PG++ P E++DE
Sbjct: 1039 NGEDTSDLYLEEREAEIQQAQQRKRDVPGILKPDEVEDE 1077
>sptr|Q7Z2B9|Q7Z2B9 CRM1 (Fragment).
Length = 1034
Score = 167 bits (423), Expect = 4e-40
Identities = 102/357 (28%), Positives = 173/357 (48%), Gaps = 1/357 (0%)
Frame = +1
Query: 22 KNQDVIRSVLNILQTNTSVASSLGPHFFPQISLIFLDMLTVYRMYSELVSSTIAEGGPFA 201
++ + ++++L+ +S+AS+ G F ++ +I D+ YR + ++ IAEGGP A
Sbjct: 657 RDSSSMMELIHVLRVFSSIASTCGTAFIDEMGIIIWDLHGFYRTFFAAQNALIAEGGPAA 716
Query: 202 S-RTSFVKLLRSVKRETLKLIETFVDKAEDLPHVGKQFVPPMMDPVLGDYARNVPDARES 378
+ + LR K+E L + E FVD E+ V +P ++ VL DY +VP+A+E
Sbjct: 717 MVQQQDARYLRLAKKEILCIFERFVDNTEERDFVAANCMPSILTVVLEDYRDSVPEAKEP 776
Query: 379 EVLSLFATIINKYKGEMLEDVPRIFEAVFQCTLEMITKNFEDYPEHRLKFFSLLRAIGTH 558
L+L +NK + D I + F T+ MI +N ED+PE R+ F LL+A+ H
Sbjct: 777 GALALVTACVNKLTTRLSGDCAAILDHTFDTTVAMICRNTEDFPEFRVNLFKLLQALNRH 836
Query: 559 CFQALIQLSSQQLKLVIDSINWAFRHTERNIAETGLSLLLEILKKFQASGFQNQFYKTYF 738
CF A + +S++ + VI + W +HT+ E GL L L+ S +F+ ++
Sbjct: 837 CFDAFLSYASRK-EDVIAGMLWVIKHTDFATMEVGLLALDSFLENVAKSELAKEFFTSFM 895
Query: 739 LTIEQEIFAVLTDTFHKPGFKLHVSVLQHLFCAVDGLTEPLWDASSVPYQYTDNAMFVRD 918
I E+ D+ H GF LH S+L LF D P + + +
Sbjct: 896 QRIFVEVLVAAMDSLHAAGFSLHCSILIKLFSVSDMFP---------PQTPVVGRVALHE 946
Query: 919 YTIKLLGTSFPNMTVAEVTKFVDGLLSSKLDLPSFKNHIRDFLVQSKEFSAQDNKDL 1089
+ ++ L P +T + +T FV S D F+ DFL++ + + A++ L
Sbjct: 947 FLVENLSV-IPTLTPSLITDFVSFAYESYCDEAEFRRRFADFLIEMQVWGAEEENRL 1002
>sptr|Q7Z2C1|Q7Z2C1 Exportin 1.
Length = 1033
Score = 153 bits (386), Expect = 8e-36
Identities = 98/349 (28%), Positives = 161/349 (46%), Gaps = 1/349 (0%)
Frame = +1
Query: 46 VLNILQTNTSVASSLGPHFFPQISLIFLDMLTVYRMYSELVSSTIAEGGPFASRTSFVKL 225
+++IL+ +S+ASS G F ++ +I D+ +YR + ++ +A+ G A +
Sbjct: 665 LIHILRVFSSIASSCGTSFVNEMGIIIYDLQGLYRTFFSAQTALVADHGTDAMERQEARY 724
Query: 226 LRSVKRETLKLIETFVDKAEDLPHVGKQFVPPMMDPVLGDYARNVPDARESEVLSLFATI 405
LR KRE L++ E FVD E+ V +P ++ VL DY ++P +E+ L+L
Sbjct: 725 LRLAKREILRIFECFVDNTEECDFVATNCMPSILTTVLEDYRDSLPIVKEAGALALVTAC 784
Query: 406 INKYKGEMLEDVPRIFEAVFQCTLEMITKNFEDYPEHRLKFFSLLRAIGTHCFQALIQLS 585
+NK + D I + T+ MI N ED+PE R+ F LL A+ T CF + +
Sbjct: 785 VNKLGTRLAGDCAAILDHTSDTTISMICANAEDFPEFRVNLFKLLHALNTRCFSNFLSYA 844
Query: 586 SQQLKLVIDSINWAFRHTERNIAETGLSLLLEILKKFQASGFQNQFYKTYFLTIEQEIFA 765
S + VI+ + W +HT+ I ETGL L L+ S FY + I E+
Sbjct: 845 STKGD-VINGMLWVIKHTDFAIMETGLKTLDAFLENVSRSELLQPFYDAFMQQIFVEVLV 903
Query: 766 VLTDTFHKPGFKLHVSVLQHLFCAVDGLTEPLWDASSVPYQYTD-NAMFVRDYTIKLLGT 942
D+ H GF+LH S+L LF +S P + + + L T
Sbjct: 904 SAMDSLHAAGFELHCSILIKLFTV----------SSMFPVDLPKLGRNDIESFLCENLST 953
Query: 943 SFPNMTVAEVTKFVDGLLSSKLDLPSFKNHIRDFLVQSKEFSAQDNKDL 1089
+T + +F+ G D F+ DFL++ + + A++ L
Sbjct: 954 -IATLTPVLIKQFIAGAYEKYGDPVEFRRSFADFLIEMQVWGAEEENRL 1001
>sptr|Q7Z2C0|Q7Z2C0 CRM1.
Length = 1034
Score = 153 bits (386), Expect = 8e-36
Identities = 100/350 (28%), Positives = 164/350 (46%), Gaps = 2/350 (0%)
Frame = +1
Query: 46 VLNILQTNTSVASSLGPHFFPQISLIFLDMLTVYRMYSELVSSTIAEGGPFASRTSFVKL 225
+++IL+ +S+ASS G F ++ +I D+ +YR + ++ +A+ G A +
Sbjct: 665 LIHILRVFSSIASSCGTSFVNEMGIIIYDLQGLYRTFFSAQTALVADHGTDAMERQEARY 724
Query: 226 LRSVKRETLKLIETFVDKAEDLPHVGKQFVPPMMDPVLGDYARNVPDARESEVLSLFATI 405
LR KRE L++ E FVD E+ V +P ++ VL DY ++P +E+ + L
Sbjct: 725 LRLAKREILRIFECFVDNTEECDFVATNCMPSILTTVLEDYRDSLPIVKEAGAIDLVTAG 784
Query: 406 INKYKGEML-EDVPRIFEAVFQCTLEMITKNFEDYPEHRLKFFSLLRAIGTHCFQALIQL 582
N+ +L ED I++ F T+ MI N ED+PE R+ F LL A+ T CF +
Sbjct: 785 FNQVGNAVLAEDCAAIWDHTFDTTISMICANAEDFPEFRVNLFKLLHALNTRCFSNFLSY 844
Query: 583 SSQQLKLVIDSINWAFRHTERNIAETGLSLLLEILKKFQASGFQNQFYKTYFLTIEQEIF 762
+S + VI+ + W +HT+ I ETGL L L+ S FY + I E+
Sbjct: 845 ASTKGD-VINGMLWVIKHTDFAIMETGLKTLDAFLENVSRSELLQPFYDAFMQQIFVEVL 903
Query: 763 AVLTDTFHKPGFKLHVSVLQHLFCAVDGLTEPLWDASSVPYQYTD-NAMFVRDYTIKLLG 939
D+ H GF+LH S+L LF +S P + + + L
Sbjct: 904 VSAMDSLHAAGFELHCSILIKLFTV----------SSMFPVDLPKLGRNDIESFLCENLS 953
Query: 940 TSFPNMTVAEVTKFVDGLLSSKLDLPSFKNHIRDFLVQSKEFSAQDNKDL 1089
T +T A + +F+ G D F+ DFL++ + + A++ L
Sbjct: 954 T-IATLTPALIKQFIAGAYEKYGDPVEFRRSFADFLIEMQVWGAEEENRL 1002
>sptr|Q7RP59|Q7RP59 Plasmodium vivax PV1H14065_P-related.
Length = 574
Score = 149 bits (376), Expect = 1e-34
Identities = 98/404 (24%), Positives = 189/404 (46%), Gaps = 45/404 (11%)
Frame = +1
Query: 7 SIDILKNQDVIRSVLNILQTNTSVASSLGPHFFPQISLIFLDMLTVYRMYSELVSSTIAE 186
+++ L N + + ++ ++ N +A +L ++ Q++L+F D L +Y++YS+ ++ +
Sbjct: 134 NLEHLCNYENSKLIITFVRVNCRLAYALSYFYYEQLNLVFFDFLKIYQLYSKYINMEVEA 193
Query: 187 GGPFASRTSFVKLLRSVKRETLKLIETFVDKA---------------------------- 282
G + + + L +KRE L LIET ++++
Sbjct: 194 NGTKRIKHAQFRNLFLMKREFLHLIETTIERSCYNIQELEEALLKREQKKMKNEIDESID 253
Query: 283 ------EDLPHVGKQFVPP----MMDPVLGDYARNVPDARESEVLSLFATIINKYKGEML 432
E+ + Q +++ +L DY + P +++EV SL +T+ K +
Sbjct: 254 VHLPTVEEAKQINFQMTSNILNVLLETILVDYRDSNPHIKDAEVFSLLSTVFKKIENVTC 313
Query: 433 EDVPRIFEAVFQCTLEMITKNFEDYPEHRLKFFSLLRAIGTHCFQALIQLSSQQLKLVID 612
+P + V T++MI +F YPEHR KF++ L A HCF L L S+ I
Sbjct: 314 PILPTVLNYVLLPTIDMIKNDFSSYPEHREKFYNFLDACVRHCFDYLFTLDSEIFNTFIQ 373
Query: 613 SINWAFRHTERNIAETGLSLLLEILKK--FQASGFQNQFYKTYFLTIEQEIFAVLTDTFH 786
S+ WA +H ++A+ GL + + L + + ++F K ++ I E+F LTD+FH
Sbjct: 374 SLLWAIKHEHPSVADHGLKITHQFLHNVIVKKKEYLDEFCKAFYYIILNEVFKTLTDSFH 433
Query: 787 KPGFKLHVSVLQHLFCAVDGLTEPLWDASSVPYQYTDNAMFVRDYTIK----LLGTSFPN 954
K GF +L +L ++ ++ ++P A + + IK L SF N
Sbjct: 434 KSGFHYQTIILMNLLRLLE------FEVINIP-----EAEITKPHIIKHVQTFLTQSFEN 482
Query: 955 MTVAEVTKFVDGLLSSKLDLP-SFKNHIRDFLVQSKEFSAQDNK 1083
+ ++ F + + ++ P +F++ +RD L+ KEFS ++
Sbjct: 483 LNQKQIETFSVDMFNFCVESPLTFRSFVRDLLISLKEFSTNQDE 526
>sptr|Q962M1|Q962M1 PV1H14065_P.
Length = 1247
Score = 149 bits (376), Expect = 1e-34
Identities = 97/391 (24%), Positives = 183/391 (46%), Gaps = 45/391 (11%)
Frame = +1
Query: 46 VLNILQTNTSVASSLGPHFFPQISLIFLDMLTVYRMYSELVSSTIAEGGPFASRTSFVKL 225
++ ++ N +A +L ++ Q++L+FLD L +Y++YS+ ++ + G + + +
Sbjct: 820 IITFVRVNCRLAYALSYFYYEQLNLVFLDFLKIYQLYSKFINMEVETNGTKRIKHAQFRN 879
Query: 226 LRSVKRETLKLIETFVDKA----------------------------------EDLPHVG 303
L +KRE L LIET ++++ E+ +
Sbjct: 880 LFLMKREFLHLIETTIERSCYNIQELEAELLKREQKKMKNEIDESMDVHLPTVEEAKQIN 939
Query: 304 KQFVPP----MMDPVLGDYARNVPDARESEVLSLFATIINKYKGEMLEDVPRIFEAVFQC 471
Q +++ +L DY + P +++EV SL +T+ K + +P + V
Sbjct: 940 FQMTSNILNVLLETILVDYRDSNPHIKDAEVFSLLSTVFKKIENVTCPILPTVLNYVLLP 999
Query: 472 TLEMITKNFEDYPEHRLKFFSLLRAIGTHCFQALIQLSSQQLKLVIDSINWAFRHTERNI 651
T++MI +F YPEHR KF++ L A HCF L L S+ I S+ WA +H ++
Sbjct: 1000 TIDMIKNDFSSYPEHREKFYNFLDACVRHCFDYLFTLDSEIFNTFIQSLLWAIKHEHPSV 1059
Query: 652 AETGLSLLLEILKK--FQASGFQNQFYKTYFLTIEQEIFAVLTDTFHKPGFKLHVSVLQH 825
A+ GL + + L + + +F K ++ I E+F LTD+FHK GF +L +
Sbjct: 1060 ADHGLRITHQFLHNIIIKKKEYLEEFCKAFYYIILNEVFKTLTDSFHKSGFHYQTIILMN 1119
Query: 826 LFCAVDGLTEPLWDASSVPYQYTDNAMFVRDYTIK----LLGTSFPNMTVAEVTKFVDGL 993
+ ++ ++ ++P +A + + +K L SF N+ ++ F L
Sbjct: 1120 MLRLLE------FEVVNLP-----DAEITKPHIVKHVQTFLTQSFENLNQKQIETFSVDL 1168
Query: 994 LSSKLDLPS-FKNHIRDFLVQSKEFSAQDNK 1083
+ ++ PS F++ +RD L+ KEFS ++
Sbjct: 1169 FNFCVESPSAFRSFVRDLLISLKEFSTNQDE 1199
>sptr|O77312|O77312 PFC0135c, MAL3P1.9 protein.
Length = 1254
Score = 147 bits (370), Expect = 6e-34
Identities = 97/390 (24%), Positives = 181/390 (46%), Gaps = 41/390 (10%)
Frame = +1
Query: 46 VLNILQTNTSVASSLGPHFFPQISLIFLDMLTVYRMYSELVSSTIAEGGPFASRTSFVKL 225
++ ++ N +A +L ++ Q++L+FLD L +Y++YS+ ++ + G + + +
Sbjct: 827 IITFVRVNCRLAYALSYFYYEQLALVFLDFLKIYQLYSKFINLEVEANGTKRIKHAQFRN 886
Query: 226 LRSVKRETLKLIETFVDKA----------------------------------EDLPHVG 303
L +KRE L LIET ++++ E+ +
Sbjct: 887 LFLMKREFLHLIETTIERSCYNIQDLEKELLKREQKKLKNEIDESMEIHLPTIEEAKQIN 946
Query: 304 KQFVPP----MMDPVLGDYARNVPDARESEVLSLFATIINKYKGEMLEDVPRIFEAVFQC 471
Q +++ +L DY + P +++EV SL +T+ K + +P + V
Sbjct: 947 FQMTSNILNVLLETILVDYRDSNPHIKDAEVFSLLSTVFKKIENVTCPILPTVLNYVLLP 1006
Query: 472 TLEMITKNFEDYPEHRLKFFSLLRAIGTHCFQALIQLSSQQLKLVIDSINWAFRHTERNI 651
T++MI +F YPEHR KF++ L A HCF L L S+ I S+ WA +H ++
Sbjct: 1007 TIDMIKNDFSSYPEHREKFYNFLDACVRHCFDYLFTLDSEIFNTFIQSLLWAIKHEHPSV 1066
Query: 652 AETGLSLLLEILKK--FQASGFQNQFYKTYFLTIEQEIFAVLTDTFHKPGFKLHVSVLQH 825
A+ GL + + L + + +F K ++ I EI LTD+FHK GF +L +
Sbjct: 1067 ADHGLRITQQFLHNIIIKKKEYLEEFCKAFYYIILNEILKTLTDSFHKSGFHYQTIILMN 1126
Query: 826 LFCAVDGLTEPLWDASSVPYQYTDNAMFVRDYTIKLLGTSFPNMTVAEVTKFVDGLLSSK 1005
L ++ ++ ++P ++ + L SF N+ ++ F + +
Sbjct: 1127 LLRLLE------FEVVNIPEVEITKPHIIK-HVQNFLTQSFENLNQKQIETFSVDMFNFC 1179
Query: 1006 LDLPS-FKNHIRDFLVQSKEFSAQDNKDLY 1092
++ PS F++ +RD L+ KEF A + +LY
Sbjct: 1180 VESPSAFRSFVRDLLISLKEF-ATNQDELY 1208
>sptr|Q8BYY5|Q8BYY5 CRM1 protein (Fragment).
Length = 193
Score = 145 bits (365), Expect = 2e-33
Identities = 77/197 (39%), Positives = 115/197 (58%), Gaps = 7/197 (3%)
Frame = +1
Query: 622 WAFRHTERNIAETGLSLLLEILKKF-QASGFQNQFYKTYFLTIEQEIFAVLTDTFHKPGF 798
WAF+HT RN+A+TGL +L +L+ Q FY+TYF I Q IF+V+TDT H G
Sbjct: 2 WAFKHTMRNVADTGLQILFTLLQNVAQEEAAAQSFYQTYFCDILQHIFSVVTDTSHTAGL 61
Query: 799 KLHVSVLQHLFCAVDG--LTEPLWDASSVPYQYTDNAMFVRDYTIKLLGTSFPNMTVAEV 972
+H S+L ++F V+ ++ PL + V +N MF++DY LL ++FP++ A+V
Sbjct: 62 TMHASILAYMFNLVEEGKISTPLNPGNPV-----NNQMFIQDYVANLLKSAFPHLQDAQV 116
Query: 973 TKFVDGLLSSKLDLPSFKNHIRDFLVQSKEFSAQDNKDLY----XXXXXXXXXXXXXXML 1140
FV GL S D+P+FK H+RDFLVQ KEF+ +D DL+ +
Sbjct: 117 KLFVTGLFSLNQDIPAFKEHLRDFLVQIKEFAGEDTSDLFLEERETALRQAQEEKHKLQM 176
Query: 1141 AIPGLIAPSELQDEMVD 1191
++PG++ P E+ +EM D
Sbjct: 177 SVPGILNPHEIPEEMCD 193
Database: swall
Posted date: Feb 26, 2004 3:00 PM
Number of letters in database: 439,479,560
Number of sequences in database: 1,381,838
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,279,880,591
Number of Sequences: 1381838
Number of extensions: 27754628
Number of successful extensions: 75543
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 70427
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 75418
length of database: 439,479,560
effective HSP length: 128
effective length of database: 262,604,296
effective search space used: 106092135584
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

