BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= QAE23e10.yg.3.1
(2083 letters)
Database: swall
1,381,838 sequences; 439,479,560 total letters
Searching..................................................done
Score E
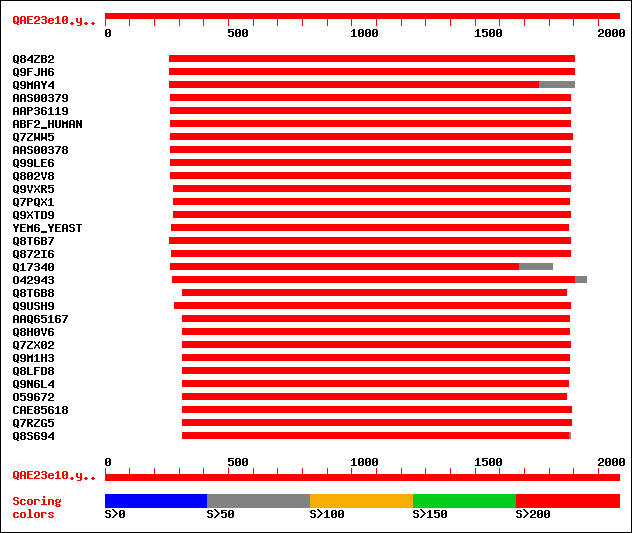
Sequences producing significant alignments: (bits) Value
sptr|Q84ZB2|Q84ZB2 Putative ABC transporter. 1016 0.0
sptr|Q9FJH6|Q9FJH6 ABC transporter homolog PnATH-like protein (A... 941 0.0
sptr|Q9MAY4|Q9MAY4 ABC transporter homolog. 885 0.0
sptrnew|AAS00379|AAS00379 Hypothetical protein ABCF2. 712 0.0
sptrnew|AAP36119|AAP36119 ATP-binding cassette, sub-family F (GC... 712 0.0
sw|Q9UG63|ABF2_HUMAN ATP-binding cassette, sub-family F, member ... 712 0.0
sptr|Q7ZWW5|Q7ZWW5 Similar to ATP-binding cassette, sub-family F... 712 0.0
sptrnew|AAS00378|AAS00378 Hypothetical protein ABCF2. 712 0.0
sptr|Q99LE6|Q99LE6 ATP-binding cassette, sub-family F, member 2. 711 0.0
sptr|Q802V8|Q802V8 Similar to ATP-binding cassette, sub-family F... 708 0.0
sptr|Q9VXR5|Q9VXR5 CG9281 protein (LD02975P). 700 0.0
sptr|Q7PQX1|Q7PQX1 ENSANGP00000010790 (Fragment). 690 0.0
sptr|Q9XTD9|Q9XTD9 T27E9.7 protein. 660 0.0
sw|P40024|YEM6_YEAST Probable ATP-dependent transporter YER036C. 570 e-161
sptr|Q8T6B7|Q8T6B7 Non-transporter ABC protein AbcF2. 566 e-160
sptr|Q872I6|Q872I6 Probable iron inhibited ABC transporter 2 (Hy... 559 e-158
sptr|Q17340|Q17340 Probable ATP-dependent transporter GNC20-2 (F... 554 e-156
sptr|O42943|O42943 Non transporter with ABC binding cassette. 521 e-146
sptr|Q8T6B8|Q8T6B8 Non-transporter ABC protein AbcF1. 463 e-129
sptr|Q9USH9|Q9USH9 Putative ABC transporter. 446 e-124
sptrnew|AAQ65167|AAQ65167 At1g64550. 425 e-117
sptr|Q8H0V6|Q8H0V6 ABC transporter protein, putative. 425 e-117
sptr|Q7ZX02|Q7ZX02 Hypothetical protein (Fragment). 417 e-115
sptr|Q9M1H3|Q9M1H3 ABC transporter-like protein (At3g54540). 415 e-114
sptr|Q8LFD8|Q8LFD8 Putative ABC transporter. 415 e-114
sptr|Q9N6L4|Q9N6L4 ABC transporter protein 1. 412 e-113
sptr|O59672|O59672 Putative amino acid starvation response, yeas... 410 e-113
sptrnew|CAE85618|CAE85618 Probable positive effector protein GCN20. 408 e-112
sptr|Q7RZG5|Q7RZG5 Hypothetical protein. 408 e-112
sptr|Q8S694|Q8S694 Putative ABC transporter. 408 e-112
>sptr|Q84ZB2|Q84ZB2 Putative ABC transporter.
Length = 592
Score = 1016 bits (2628), Expect = 0.0
Identities = 502/547 (91%), Positives = 525/547 (95%)
Frame = +1
Query: 262 LKLSDRTCTGVLASHPLSRDIHIESLSLTFHGHDLIVDSDLEXXXXXXXXXXXXXXCGKS 441
LKLSDRTCTGVLASHPLSRDIHIESLSLTFHGHDLIVDS+LE CGKS
Sbjct: 46 LKLSDRTCTGVLASHPLSRDIHIESLSLTFHGHDLIVDSELELNYGRRYGLLGLNGCGKS 105
Query: 442 TLLTAIGCRELPIPEHMDIYHLSHEIEASDMSALQAIVSCDEERVKLEKEAEILAAQDDG 621
TLLTAIGCRELPIPEHMDIYHLS EIEASDMSALQA++ CDEER+KLEKEAEIL+AQDDG
Sbjct: 106 TLLTAIGCRELPIPEHMDIYHLSSEIEASDMSALQAVICCDEERMKLEKEAEILSAQDDG 165
Query: 622 GGEALERVYERLDAMDAGTAEKRAAEILFGLGFDKQMQAKKTKDFSGGWRMRIALARALF 801
GG+AL+R+YERL+A+DA TAEKRAAEILFGLGF+KQMQAKKT+DFSGGWRMRIALARALF
Sbjct: 166 GGDALDRIYERLEALDASTAEKRAAEILFGLGFNKQMQAKKTQDFSGGWRMRIALARALF 225
Query: 802 MNPTILLLDEPTNHLDLEACVWLEETLKKFDRILVVISHSQDFLNGVCTNIIHMQNKKLK 981
MNPTILLLDEPTNHLDLEACVWLEETLKKFDRILVVISHSQDFLNGVCTNIIHMQ+KKLK
Sbjct: 226 MNPTILLLDEPTNHLDLEACVWLEETLKKFDRILVVISHSQDFLNGVCTNIIHMQSKKLK 285
Query: 982 LYTGNYDQYVQTRSDLEENQMKQYKWEQEQIASMKEYIARFGHGSAKLARQAQSKEKTLA 1161
LY+GNYDQYVQTRS+LEENQMKQYKWEQEQIASMKEYIARFGHGSAKLARQAQSKEKTLA
Sbjct: 286 LYSGNYDQYVQTRSELEENQMKQYKWEQEQIASMKEYIARFGHGSAKLARQAQSKEKTLA 345
Query: 1162 KMERGGLTEKVVRDRVLVFRFTDVGKLPPPVLQFVEVKFGYTPDNLIYKSLDFGVDLDSR 1341
KMERGGLTEKVVRD+VLVFRFTDVGKLPPPVLQFVEV FGYTPDNLIYK+LDFGVDLDSR
Sbjct: 346 KMERGGLTEKVVRDKVLVFRFTDVGKLPPPVLQFVEVSFGYTPDNLIYKNLDFGVDLDSR 405
Query: 1342 IALVGPNGAGKSTLLKLMTGDLAPLDGMVRRHNHLRIAQYHQHLADKLDLDMPALAYMMK 1521
IALVGPNGAGKSTLLKLMTGDLAPLDGMVRRHNHLRIAQYHQHLA+KLDLDMPAL YMM+
Sbjct: 406 IALVGPNGAGKSTLLKLMTGDLAPLDGMVRRHNHLRIAQYHQHLAEKLDLDMPALQYMMR 465
Query: 1522 EYPGTEEEKMRAAVGRFGLSGKAQVMPMRNLSDGQRSRVIFAWLAYRQPQLLLLDEPTNH 1701
EYPG EEEKMRAA+G+FGLSGKAQVMPMRNLSDGQRSRVIFAWLAYRQPQLLLLDEPTNH
Sbjct: 466 EYPGNEEEKMRAAIGKFGLSGKAQVMPMRNLSDGQRSRVIFAWLAYRQPQLLLLDEPTNH 525
Query: 1702 LDIETIDSLAEALNEWDGGLVLVSHDFRLINQVAQEIWVCENQAVTRWEGDIMDFKAHLK 1881
LDIETIDSLAEALNEWDGGLVLVSHDFRLINQVAQEIWVCE QAVTRWEGDIMDFK HL+
Sbjct: 526 LDIETIDSLAEALNEWDGGLVLVSHDFRLINQVAQEIWVCEKQAVTRWEGDIMDFKEHLR 585
Query: 1882 SKAGLSD 1902
S+AGLSD
Sbjct: 586 SRAGLSD 592
>sptr|Q9FJH6|Q9FJH6 ABC transporter homolog PnATH-like protein
(At5g60790).
Length = 595
Score = 941 bits (2431), Expect = 0.0
Identities = 458/547 (83%), Positives = 501/547 (91%)
Frame = +1
Query: 262 LKLSDRTCTGVLASHPLSRDIHIESLSLTFHGHDLIVDSDLEXXXXXXXXXXXXXXCGKS 441
L++SDRTCTGVL SHP SRDI IESLS+TFHG+DLIVDS LE CGKS
Sbjct: 49 LQISDRTCTGVLCSHPQSRDIRIESLSVTFHGYDLIVDSMLELNYGRRYGLLGLNGCGKS 108
Query: 442 TLLTAIGCRELPIPEHMDIYHLSHEIEASDMSALQAIVSCDEERVKLEKEAEILAAQDDG 621
TLLTAIG RE+PIP+ MDIYHLSHEIEA+DMS+L+A+VSCDEER++LEKE EIL QDDG
Sbjct: 109 TLLTAIGRREIPIPDQMDIYHLSHEIEATDMSSLEAVVSCDEERLRLEKEVEILVQQDDG 168
Query: 622 GGEALERVYERLDAMDAGTAEKRAAEILFGLGFDKQMQAKKTKDFSGGWRMRIALARALF 801
GGE L+ +YERLDAMDA TAEKRAAEILFGLGFDK+MQAKKTKDFSGGWRMRIALARALF
Sbjct: 169 GGERLQSIYERLDAMDAETAEKRAAEILFGLGFDKEMQAKKTKDFSGGWRMRIALARALF 228
Query: 802 MNPTILLLDEPTNHLDLEACVWLEETLKKFDRILVVISHSQDFLNGVCTNIIHMQNKKLK 981
+ PTILLLDEPTNHLDLEACVWLEE+LK FDRILVV+SHSQDFLNGVCTNIIHMQ+K+LK
Sbjct: 229 IMPTILLLDEPTNHLDLEACVWLEESLKNFDRILVVVSHSQDFLNGVCTNIIHMQSKQLK 288
Query: 982 LYTGNYDQYVQTRSDLEENQMKQYKWEQEQIASMKEYIARFGHGSAKLARQAQSKEKTLA 1161
YTGN+DQY QTRS+LEENQMKQY+WEQEQI+ MKEYIARFGHGSAKLARQAQSKEKTLA
Sbjct: 289 YYTGNFDQYCQTRSELEENQMKQYRWEQEQISHMKEYIARFGHGSAKLARQAQSKEKTLA 348
Query: 1162 KMERGGLTEKVVRDRVLVFRFTDVGKLPPPVLQFVEVKFGYTPDNLIYKSLDFGVDLDSR 1341
KMERGGLTEKV RD VLVFRF DVGKLPPPVLQFVEV FGYTPD LIYK++DFGVDLDSR
Sbjct: 349 KMERGGLTEKVARDSVLVFRFADVGKLPPPVLQFVEVSFGYTPDYLIYKNIDFGVDLDSR 408
Query: 1342 IALVGPNGAGKSTLLKLMTGDLAPLDGMVRRHNHLRIAQYHQHLADKLDLDMPALAYMMK 1521
+ALVGPNGAGKSTLLKLMTG+L P +GMVRRHNHL+IAQYHQHLA+KLDL++PAL YMM+
Sbjct: 409 VALVGPNGAGKSTLLKLMTGELHPTEGMVRRHNHLKIAQYHQHLAEKLDLELPALLYMMR 468
Query: 1522 EYPGTEEEKMRAAVGRFGLSGKAQVMPMRNLSDGQRSRVIFAWLAYRQPQLLLLDEPTNH 1701
E+PGTEEEKMRAA+GRFGL+GKAQVMPM+NLSDGQRSRVIFAWLAY+QP +LLLDEPTNH
Sbjct: 469 EFPGTEEEKMRAAIGRFGLTGKAQVMPMKNLSDGQRSRVIFAWLAYKQPNMLLLDEPTNH 528
Query: 1702 LDIETIDSLAEALNEWDGGLVLVSHDFRLINQVAQEIWVCENQAVTRWEGDIMDFKAHLK 1881
LDIETIDSLAEALNEWDGGLVLVSHDFRLINQVA EIWVCE Q +T+W GDIMDFK HLK
Sbjct: 529 LDIETIDSLAEALNEWDGGLVLVSHDFRLINQVAHEIWVCEKQCITKWNGDIMDFKRHLK 588
Query: 1882 SKAGLSD 1902
+KAGL D
Sbjct: 589 AKAGLED 595
>sptr|Q9MAY4|Q9MAY4 ABC transporter homolog.
Length = 565
Score = 885 bits (2288), Expect = 0.0
Identities = 431/498 (86%), Positives = 469/498 (94%)
Frame = +1
Query: 262 LKLSDRTCTGVLASHPLSRDIHIESLSLTFHGHDLIVDSDLEXXXXXXXXXXXXXXCGKS 441
L++SDRTCTGVL SHPLSRDI IESLS+TFHGHDLIVDS+LE CGKS
Sbjct: 57 LQISDRTCTGVLCSHPLSRDIRIESLSVTFHGHDLIVDSELELNYGRRYGLLGLNGCGKS 116
Query: 442 TLLTAIGCRELPIPEHMDIYHLSHEIEASDMSALQAIVSCDEERVKLEKEAEILAAQDDG 621
TLL AIGCRELPIP+HMDIYHL+ EIEASDMS+L+A++SCDEER+KLEKEAE+LAA+DDG
Sbjct: 117 TLLAAIGCRELPIPDHMDIYHLTREIEASDMSSLEAVISCDEERLKLEKEAEVLAAKDDG 176
Query: 622 GGEALERVYERLDAMDAGTAEKRAAEILFGLGFDKQMQAKKTKDFSGGWRMRIALARALF 801
GGEAL+R+YERL+AMDA TAEKRAAEIL+GLGF+K+MQ KKT+DFSGGWRMRIALARALF
Sbjct: 177 GGEALDRIYERLEAMDASTAEKRAAEILYGLGFNKKMQEKKTRDFSGGWRMRIALARALF 236
Query: 802 MNPTILLLDEPTNHLDLEACVWLEETLKKFDRILVVISHSQDFLNGVCTNIIHMQNKKLK 981
MNPTILLLDEPTNHLDLEACVWLEETLKKFDRILVV+SHSQDFLNGVCTNIIHMQNKKLK
Sbjct: 237 MNPTILLLDEPTNHLDLEACVWLEETLKKFDRILVVVSHSQDFLNGVCTNIIHMQNKKLK 296
Query: 982 LYTGNYDQYVQTRSDLEENQMKQYKWEQEQIASMKEYIARFGHGSAKLARQAQSKEKTLA 1161
+YTGN+DQYVQTRS+LEENQMKQYKWEQ+QIASMKEYIARFGHGSAKLARQAQSKEKTLA
Sbjct: 297 IYTGNFDQYVQTRSELEENQMKQYKWEQDQIASMKEYIARFGHGSAKLARQAQSKEKTLA 356
Query: 1162 KMERGGLTEKVVRDRVLVFRFTDVGKLPPPVLQFVEVKFGYTPDNLIYKSLDFGVDLDSR 1341
KMERGGLTEKV RD++LVFRF +VGKLPPPVLQFVEV FGYTPDNLIYKS+DFGVDLDSR
Sbjct: 357 KMERGGLTEKVARDKILVFRFVNVGKLPPPVLQFVEVTFGYTPDNLIYKSIDFGVDLDSR 416
Query: 1342 IALVGPNGAGKSTLLKLMTGDLAPLDGMVRRHNHLRIAQYHQHLADKLDLDMPALAYMMK 1521
IALVGPNGAGKSTLLKLMTGDL PLDGMVRRHNHLRIAQ+HQHLA+KLDL+M AL YM+K
Sbjct: 417 IALVGPNGAGKSTLLKLMTGDLVPLDGMVRRHNHLRIAQFHQHLAEKLDLEMSALQYMIK 476
Query: 1522 EYPGTEEEKMRAAVGRFGLSGKAQVMPMRNLSDGQRSRVIFAWLAYRQPQLLLLDEPTNH 1701
EYPG EEE+MRAA+G+FGL+GKAQVMPM+NLSDGQRSRVIFAWLAYRQP LLLLDEPTNH
Sbjct: 477 EYPGNEEERMRAAIGKFGLTGKAQVMPMKNLSDGQRSRVIFAWLAYRQPHLLLLDEPTNH 536
Query: 1702 LDIETIDSLAEALNEWDG 1755
LDIETIDSLAEALNEW G
Sbjct: 537 LDIETIDSLAEALNEWMG 554
Score = 73.9 bits (180), Expect = 9e-12
Identities = 64/214 (29%), Positives = 100/214 (46%), Gaps = 26/214 (12%)
Frame = +1
Query: 1339 RIALVGPNGAGKSTLL---------------------KLMTGDLAPLDGMVR-RHNHLRI 1452
R L+G NG GKSTLL ++ D++ L+ ++ L++
Sbjct: 104 RYGLLGLNGCGKSTLLAAIGCRELPIPDHMDIYHLTREIEASDMSSLEAVISCDEERLKL 163
Query: 1453 AQYHQHLADKLDLDMPAL--AYMMKEYPGTEEEKMRAAVGRFGL--SGKAQVMPMRNLSD 1620
+ + LA K D AL Y E + RAA +GL + K Q R+ S
Sbjct: 164 EKEAEVLAAKDDGGGEALDRIYERLEAMDASTAEKRAAEILYGLGFNKKMQEKKTRDFSG 223
Query: 1621 GQRSRVIFAWLAYRQPQLLLLDEPTNHLDIETIDSLAEALNEWDGGLVLVSHDFRLINQV 1800
G R R+ A + P +LLLDEPTNHLD+E L E L ++D LV+VSH +N V
Sbjct: 224 GWRMRIALARALFMNPTILLLDEPTNHLDLEACVWLEETLKKFDRILVVVSHSQDFLNGV 283
Query: 1801 AQEIWVCENQAVTRWEGDIMDFKAHLKSKAGLSD 1902
I +N+ + + G +F ++++++ L +
Sbjct: 284 CTNIIHMQNKKLKIYTG---NFDQYVQTRSELEE 314
>sptrnew|AAS00379|AAS00379 Hypothetical protein ABCF2.
Length = 623
Score = 712 bits (1838), Expect = 0.0
Identities = 353/543 (65%), Positives = 424/543 (78%), Gaps = 2/543 (0%)
Frame = +1
Query: 265 KLSDRTCTGVLASHPLSRDIHIESLSLTFHGHDLIVDSDLEXXXXXXXXXXXXXXCGKST 444
K + R TGVLASHP S D+HI +LSLTFHG +L+ D+ LE GKS
Sbjct: 67 KAAARAVTGVLASHPNSTDVHIINLSLTFHGQELLSDTKLELNSGRRYGLIGLNGIGKSM 126
Query: 445 LLTAIGCRELPIPEHMDIYHLSHEIEASDMSALQAIVSCDEERVKLEKEAEILAAQDDGG 624
LL+AIG RE+PIPEH+DIYHL+ E+ SD + L ++ D ER LEKEAE LA +D
Sbjct: 127 LLSAIGKREVPIPEHIDIYHLTREMPPSDKTPLHCVMEVDTERAMLEKEAERLA-HEDAE 185
Query: 625 GEALERVYERLDAMDAGTAEKRAAEILFGLGFDKQMQAKKTKDFSGGWRMRIALARALFM 804
E L +YERL+ +DA AE RA+ IL GLGF MQ KK KDFSGGWRMR+ALARALF+
Sbjct: 186 CEKLMELYERLEELDADKAEMRASRILHGLGFTPAMQRKKLKDFSGGWRMRVALARALFI 245
Query: 805 NPTILLLDEPTNHLDLEACVWLEETLKKFDRILVVISHSQDFLNGVCTNIIHMQNKKLKL 984
P +LLLDEPTNHLDL+ACVWLEE LK F RILV++SHSQDFLNGVCTNIIHM NKKLK
Sbjct: 246 RPFMLLLDEPTNHLDLDACVWLEEELKTFKRILVLVSHSQDFLNGVCTNIIHMHNKKLKY 305
Query: 985 YTGNYDQYVQTRSDLEENQMKQYKWEQEQIASMKEYIARFGHGSAKLARQAQSKEKTLAK 1164
YTGNYDQYV+TR +LEENQMK++ WEQ+QIA MK YIARFGHGSAKLARQAQSKEKTL K
Sbjct: 306 YTGNYDQYVKTRLELEENQMKRFHWEQDQIAHMKNYIARFGHGSAKLARQAQSKEKTLQK 365
Query: 1165 MERGGLTEKVVRDRVLVFRFTDVGKLPPPVLQFVEVKFGYTPDN-LIYKSLDFGVDLDSR 1341
M GLTE+VV D+ L F F GK+PPPV+ V F YT D IY +L+FG+DLD+R
Sbjct: 366 MMASGLTERVVSDKTLSFYFPPCGKIPPPVIMVQNVSFKYTKDGPCIYNNLEFGIDLDTR 425
Query: 1342 IALVGPNGAGKSTLLKLMTGDLAPLDGMVRRHNHLRIAQYHQHLADKLDLDMPALAYMMK 1521
+ALVGPNGAGKSTLLKL+TG+L P DGM+R+H+H++I +YHQHL ++LDLD+ L YMMK
Sbjct: 426 VALVGPNGAGKSTLLKLLTGELLPTDGMIRKHSHVKIGRYHQHLQEQLDLDLSPLEYMMK 485
Query: 1522 EYPG-TEEEKMRAAVGRFGLSGKAQVMPMRNLSDGQRSRVIFAWLAYRQPQLLLLDEPTN 1698
YP E+E+MR +GR+GL+GK QV P+RNLSDGQ+ RV AWLA++ P +L LDEPTN
Sbjct: 486 CYPEIKEKEEMRKIIGRYGLTGKQQVSPIRNLSDGQKCRVCLAWLAWQNPHMLFLDEPTN 545
Query: 1699 HLDIETIDSLAEALNEWDGGLVLVSHDFRLINQVAQEIWVCENQAVTRWEGDIMDFKAHL 1878
HLDIETID+LA+A+NE++GG++LVSHDFRLI QVAQEIWVCE Q +T+W GDI+ +K HL
Sbjct: 546 HLDIETIDALADAINEFEGGMMLVSHDFRLIQQVAQEIWVCEKQTITKWPGDILAYKEHL 605
Query: 1879 KSK 1887
KSK
Sbjct: 606 KSK 608
>sptrnew|AAP36119|AAP36119 ATP-binding cassette, sub-family F (GCN20),
member 2.
Length = 623
Score = 712 bits (1838), Expect = 0.0
Identities = 353/543 (65%), Positives = 424/543 (78%), Gaps = 2/543 (0%)
Frame = +1
Query: 265 KLSDRTCTGVLASHPLSRDIHIESLSLTFHGHDLIVDSDLEXXXXXXXXXXXXXXCGKST 444
K + R TGVLASHP S D+HI +LSLTFHG +L+ D+ LE GKS
Sbjct: 67 KAAARAVTGVLASHPNSTDVHIINLSLTFHGQELLSDTKLELNSGRRYGLIGLNGIGKSM 126
Query: 445 LLTAIGCRELPIPEHMDIYHLSHEIEASDMSALQAIVSCDEERVKLEKEAEILAAQDDGG 624
LL+AIG RE+PIPEH+DIYHL+ E+ SD + L ++ D ER LEKEAE LA +D
Sbjct: 127 LLSAIGKREVPIPEHIDIYHLTREMPPSDKTPLHCVMEVDTERAMLEKEAERLA-HEDAE 185
Query: 625 GEALERVYERLDAMDAGTAEKRAAEILFGLGFDKQMQAKKTKDFSGGWRMRIALARALFM 804
E L +YERL+ +DA AE RA+ IL GLGF MQ KK KDFSGGWRMR+ALARALF+
Sbjct: 186 CEKLMELYERLEELDADKAEMRASRILHGLGFTPAMQRKKLKDFSGGWRMRVALARALFI 245
Query: 805 NPTILLLDEPTNHLDLEACVWLEETLKKFDRILVVISHSQDFLNGVCTNIIHMQNKKLKL 984
P +LLLDEPTNHLDL+ACVWLEE LK F RILV++SHSQDFLNGVCTNIIHM NKKLK
Sbjct: 246 RPFMLLLDEPTNHLDLDACVWLEEELKTFKRILVLVSHSQDFLNGVCTNIIHMHNKKLKY 305
Query: 985 YTGNYDQYVQTRSDLEENQMKQYKWEQEQIASMKEYIARFGHGSAKLARQAQSKEKTLAK 1164
YTGNYDQYV+TR +LEENQMK++ WEQ+QIA MK YIARFGHGSAKLARQAQSKEKTL K
Sbjct: 306 YTGNYDQYVKTRLELEENQMKRFHWEQDQIAHMKNYIARFGHGSAKLARQAQSKEKTLQK 365
Query: 1165 MERGGLTEKVVRDRVLVFRFTDVGKLPPPVLQFVEVKFGYTPDN-LIYKSLDFGVDLDSR 1341
M GLTE+VV D+ L F F GK+PPPV+ V F YT D IY +L+FG+DLD+R
Sbjct: 366 MMASGLTERVVSDKTLSFYFPPCGKIPPPVIMVQNVSFKYTKDGPCIYNNLEFGIDLDTR 425
Query: 1342 IALVGPNGAGKSTLLKLMTGDLAPLDGMVRRHNHLRIAQYHQHLADKLDLDMPALAYMMK 1521
+ALVGPNGAGKSTLLKL+TG+L P DGM+R+H+H++I +YHQHL ++LDLD+ L YMMK
Sbjct: 426 VALVGPNGAGKSTLLKLLTGELLPTDGMIRKHSHVKIGRYHQHLQEQLDLDLSPLEYMMK 485
Query: 1522 EYPG-TEEEKMRAAVGRFGLSGKAQVMPMRNLSDGQRSRVIFAWLAYRQPQLLLLDEPTN 1698
YP E+E+MR +GR+GL+GK QV P+RNLSDGQ+ RV AWLA++ P +L LDEPTN
Sbjct: 486 CYPEIKEKEEMRKIIGRYGLTGKQQVSPIRNLSDGQKCRVCLAWLAWQNPHMLFLDEPTN 545
Query: 1699 HLDIETIDSLAEALNEWDGGLVLVSHDFRLINQVAQEIWVCENQAVTRWEGDIMDFKAHL 1878
HLDIETID+LA+A+NE++GG++LVSHDFRLI QVAQEIWVCE Q +T+W GDI+ +K HL
Sbjct: 546 HLDIETIDALADAINEFEGGMMLVSHDFRLIQQVAQEIWVCEKQTITKWPGDILAYKEHL 605
Query: 1879 KSK 1887
KSK
Sbjct: 606 KSK 608
>sw|Q9UG63|ABF2_HUMAN ATP-binding cassette, sub-family F, member 2
(Iron inhibited ABC transporter 2) (HUSSY-18).
Length = 623
Score = 712 bits (1838), Expect = 0.0
Identities = 353/543 (65%), Positives = 424/543 (78%), Gaps = 2/543 (0%)
Frame = +1
Query: 265 KLSDRTCTGVLASHPLSRDIHIESLSLTFHGHDLIVDSDLEXXXXXXXXXXXXXXCGKST 444
K + R TGVLASHP S D+HI +LSLTFHG +L+ D+ LE GKS
Sbjct: 67 KAAARAVTGVLASHPNSTDVHIINLSLTFHGQELLSDTKLELNSGRRYGLIGLNGIGKSM 126
Query: 445 LLTAIGCRELPIPEHMDIYHLSHEIEASDMSALQAIVSCDEERVKLEKEAEILAAQDDGG 624
LL+AIG RE+PIPEH+DIYHL+ E+ SD + L ++ D ER LEKEAE LA +D
Sbjct: 127 LLSAIGKREVPIPEHIDIYHLTREMPPSDKTPLHCVMEVDTERAMLEKEAERLA-HEDAE 185
Query: 625 GEALERVYERLDAMDAGTAEKRAAEILFGLGFDKQMQAKKTKDFSGGWRMRIALARALFM 804
E L +YERL+ +DA AE RA+ IL GLGF MQ KK KDFSGGWRMR+ALARALF+
Sbjct: 186 CEKLMELYERLEELDADKAEMRASRILHGLGFTPAMQRKKLKDFSGGWRMRVALARALFI 245
Query: 805 NPTILLLDEPTNHLDLEACVWLEETLKKFDRILVVISHSQDFLNGVCTNIIHMQNKKLKL 984
P +LLLDEPTNHLDL+ACVWLEE LK F RILV++SHSQDFLNGVCTNIIHM NKKLK
Sbjct: 246 RPFMLLLDEPTNHLDLDACVWLEEELKTFKRILVLVSHSQDFLNGVCTNIIHMHNKKLKY 305
Query: 985 YTGNYDQYVQTRSDLEENQMKQYKWEQEQIASMKEYIARFGHGSAKLARQAQSKEKTLAK 1164
YTGNYDQYV+TR +LEENQMK++ WEQ+QIA MK YIARFGHGSAKLARQAQSKEKTL K
Sbjct: 306 YTGNYDQYVKTRLELEENQMKRFHWEQDQIAHMKNYIARFGHGSAKLARQAQSKEKTLQK 365
Query: 1165 MERGGLTEKVVRDRVLVFRFTDVGKLPPPVLQFVEVKFGYTPDN-LIYKSLDFGVDLDSR 1341
M GLTE+VV D+ L F F GK+PPPV+ V F YT D IY +L+FG+DLD+R
Sbjct: 366 MMASGLTERVVSDKTLSFYFPPCGKIPPPVIMVQNVSFKYTKDGPCIYNNLEFGIDLDTR 425
Query: 1342 IALVGPNGAGKSTLLKLMTGDLAPLDGMVRRHNHLRIAQYHQHLADKLDLDMPALAYMMK 1521
+ALVGPNGAGKSTLLKL+TG+L P DGM+R+H+H++I +YHQHL ++LDLD+ L YMMK
Sbjct: 426 VALVGPNGAGKSTLLKLLTGELLPTDGMIRKHSHVKIGRYHQHLQEQLDLDLSPLEYMMK 485
Query: 1522 EYPG-TEEEKMRAAVGRFGLSGKAQVMPMRNLSDGQRSRVIFAWLAYRQPQLLLLDEPTN 1698
YP E+E+MR +GR+GL+GK QV P+RNLSDGQ+ RV AWLA++ P +L LDEPTN
Sbjct: 486 CYPEIKEKEEMRKIIGRYGLTGKQQVSPIRNLSDGQKCRVCLAWLAWQNPHMLFLDEPTN 545
Query: 1699 HLDIETIDSLAEALNEWDGGLVLVSHDFRLINQVAQEIWVCENQAVTRWEGDIMDFKAHL 1878
HLDIETID+LA+A+NE++GG++LVSHDFRLI QVAQEIWVCE Q +T+W GDI+ +K HL
Sbjct: 546 HLDIETIDALADAINEFEGGMMLVSHDFRLIQQVAQEIWVCEKQTITKWPGDILAYKEHL 605
Query: 1879 KSK 1887
KSK
Sbjct: 606 KSK 608
>sptr|Q7ZWW5|Q7ZWW5 Similar to ATP-binding cassette, sub-family F
(GCN20), member 2.
Length = 607
Score = 712 bits (1838), Expect = 0.0
Identities = 351/545 (64%), Positives = 429/545 (78%), Gaps = 2/545 (0%)
Frame = +1
Query: 265 KLSDRTCTGVLASHPLSRDIHIESLSLTFHGHDLIVDSDLEXXXXXXXXXXXXXXCGKST 444
K R TGVLASHP S D+HI ++SLTFHG +L+ D+ +E GKS
Sbjct: 62 KAEARAVTGVLASHPNSTDVHIINMSLTFHGQELLSDTTIELNSGRRYGLIGLNGTGKSM 121
Query: 445 LLTAIGCRELPIPEHMDIYHLSHEIEASDMSALQAIVSCDEERVKLEKEAEILAAQDDGG 624
LL+A+G RE+PIP+H+DIYHL+ E+ SD SALQ ++ D ER LEKEAE LA +D
Sbjct: 122 LLSAVGGREVPIPDHIDIYHLTREMPPSDKSALQCVMEVDTERNMLEKEAERLA-HEDAE 180
Query: 625 GEALERVYERLDAMDAGTAEKRAAEILFGLGFDKQMQAKKTKDFSGGWRMRIALARALFM 804
E L +YERL+ +DA AE RA+ IL GLGF MQ KK KDFSGGWRMR+ALARALF+
Sbjct: 181 CEKLMELYERLEELDAAKAEVRASRILHGLGFTPAMQRKKLKDFSGGWRMRVALARALFI 240
Query: 805 NPTILLLDEPTNHLDLEACVWLEETLKKFDRILVVISHSQDFLNGVCTNIIHMQNKKLKL 984
P +LLLDEPTNHLDLEACVWLEE LK F RILV+ISHSQDFLNGVCTNIIH+ N+KLK
Sbjct: 241 RPFMLLLDEPTNHLDLEACVWLEEELKSFRRILVLISHSQDFLNGVCTNIIHLHNRKLKY 300
Query: 985 YTGNYDQYVQTRSDLEENQMKQYKWEQEQIASMKEYIARFGHGSAKLARQAQSKEKTLAK 1164
YTGNYDQYV+TR +LEENQMK++ WEQ+QIA MK YIARFGHGSAKLARQAQSKEKTL K
Sbjct: 301 YTGNYDQYVKTRLELEENQMKRFNWEQDQIAHMKNYIARFGHGSAKLARQAQSKEKTLQK 360
Query: 1165 MERGGLTEKVVRDRVLVFRFTDVGKLPPPVLQFVEVKFGYTPDN-LIYKSLDFGVDLDSR 1341
M GLTE+VV D+ L F F GK+PPPV+ V F Y+P+ LIYK+L+FG+DLD+R
Sbjct: 361 MMSSGLTERVVSDKTLSFCFPSCGKIPPPVIMVQHVSFRYSPEGPLIYKNLEFGIDLDTR 420
Query: 1342 IALVGPNGAGKSTLLKLMTGDLAPLDGMVRRHNHLRIAQYHQHLADKLDLDMPALAYMMK 1521
+ALVGPNGAGKSTLLKL+ G+L P DGM+R+++H++I +YHQHL ++L+L++ L YMMK
Sbjct: 421 VALVGPNGAGKSTLLKLLAGELLPSDGMIRKNSHVKIGRYHQHLTEQLELELSPLEYMMK 480
Query: 1522 EYPG-TEEEKMRAAVGRFGLSGKAQVMPMRNLSDGQRSRVIFAWLAYRQPQLLLLDEPTN 1698
YP E+E+MR +GR+GL+GK QV P+RNLSDGQ+ RV FAWLA++ P +L LDEPTN
Sbjct: 481 CYPDIKEKEEMRKIIGRYGLTGKQQVSPIRNLSDGQKCRVCFAWLAWQNPHMLFLDEPTN 540
Query: 1699 HLDIETIDSLAEALNEWDGGLVLVSHDFRLINQVAQEIWVCENQAVTRWEGDIMDFKAHL 1878
HLDIETID+LA+A+NE++GG++LVSHDFRLI QVAQEIWVCE QAVT+W G I+ +K HL
Sbjct: 541 HLDIETIDALADAINEFEGGMMLVSHDFRLIQQVAQEIWVCEKQAVTKWSGGILSYKQHL 600
Query: 1879 KSKAG 1893
KS+ G
Sbjct: 601 KSRLG 605
>sptrnew|AAS00378|AAS00378 Hypothetical protein ABCF2.
Length = 634
Score = 712 bits (1838), Expect = 0.0
Identities = 353/543 (65%), Positives = 424/543 (78%), Gaps = 2/543 (0%)
Frame = +1
Query: 265 KLSDRTCTGVLASHPLSRDIHIESLSLTFHGHDLIVDSDLEXXXXXXXXXXXXXXCGKST 444
K + R TGVLASHP S D+HI +LSLTFHG +L+ D+ LE GKS
Sbjct: 67 KAAARAVTGVLASHPNSTDVHIINLSLTFHGQELLSDTKLELNSGRRYGLIGLNGIGKSM 126
Query: 445 LLTAIGCRELPIPEHMDIYHLSHEIEASDMSALQAIVSCDEERVKLEKEAEILAAQDDGG 624
LL+AIG RE+PIPEH+DIYHL+ E+ SD + L ++ D ER LEKEAE LA +D
Sbjct: 127 LLSAIGKREVPIPEHIDIYHLTREMPPSDKTPLHCVMEVDTERAMLEKEAERLA-HEDAE 185
Query: 625 GEALERVYERLDAMDAGTAEKRAAEILFGLGFDKQMQAKKTKDFSGGWRMRIALARALFM 804
E L +YERL+ +DA AE RA+ IL GLGF MQ KK KDFSGGWRMR+ALARALF+
Sbjct: 186 CEKLMELYERLEELDADKAEMRASRILHGLGFTPAMQRKKLKDFSGGWRMRVALARALFI 245
Query: 805 NPTILLLDEPTNHLDLEACVWLEETLKKFDRILVVISHSQDFLNGVCTNIIHMQNKKLKL 984
P +LLLDEPTNHLDL+ACVWLEE LK F RILV++SHSQDFLNGVCTNIIHM NKKLK
Sbjct: 246 RPFMLLLDEPTNHLDLDACVWLEEELKTFKRILVLVSHSQDFLNGVCTNIIHMHNKKLKY 305
Query: 985 YTGNYDQYVQTRSDLEENQMKQYKWEQEQIASMKEYIARFGHGSAKLARQAQSKEKTLAK 1164
YTGNYDQYV+TR +LEENQMK++ WEQ+QIA MK YIARFGHGSAKLARQAQSKEKTL K
Sbjct: 306 YTGNYDQYVKTRLELEENQMKRFHWEQDQIAHMKNYIARFGHGSAKLARQAQSKEKTLQK 365
Query: 1165 MERGGLTEKVVRDRVLVFRFTDVGKLPPPVLQFVEVKFGYTPDN-LIYKSLDFGVDLDSR 1341
M GLTE+VV D+ L F F GK+PPPV+ V F YT D IY +L+FG+DLD+R
Sbjct: 366 MMASGLTERVVSDKTLSFYFPPCGKIPPPVIMVQNVSFKYTKDGPCIYNNLEFGIDLDTR 425
Query: 1342 IALVGPNGAGKSTLLKLMTGDLAPLDGMVRRHNHLRIAQYHQHLADKLDLDMPALAYMMK 1521
+ALVGPNGAGKSTLLKL+TG+L P DGM+R+H+H++I +YHQHL ++LDLD+ L YMMK
Sbjct: 426 VALVGPNGAGKSTLLKLLTGELLPTDGMIRKHSHVKIGRYHQHLQEQLDLDLSPLEYMMK 485
Query: 1522 EYPG-TEEEKMRAAVGRFGLSGKAQVMPMRNLSDGQRSRVIFAWLAYRQPQLLLLDEPTN 1698
YP E+E+MR +GR+GL+GK QV P+RNLSDGQ+ RV AWLA++ P +L LDEPTN
Sbjct: 486 CYPEIKEKEEMRKIIGRYGLTGKQQVSPIRNLSDGQKCRVCLAWLAWQNPHMLFLDEPTN 545
Query: 1699 HLDIETIDSLAEALNEWDGGLVLVSHDFRLINQVAQEIWVCENQAVTRWEGDIMDFKAHL 1878
HLDIETID+LA+A+NE++GG++LVSHDFRLI QVAQEIWVCE Q +T+W GDI+ +K HL
Sbjct: 546 HLDIETIDALADAINEFEGGMMLVSHDFRLIQQVAQEIWVCEKQTITKWPGDILAYKEHL 605
Query: 1879 KSK 1887
KSK
Sbjct: 606 KSK 608
>sptr|Q99LE6|Q99LE6 ATP-binding cassette, sub-family F, member 2.
Length = 628
Score = 711 bits (1836), Expect = 0.0
Identities = 352/543 (64%), Positives = 425/543 (78%), Gaps = 2/543 (0%)
Frame = +1
Query: 265 KLSDRTCTGVLASHPLSRDIHIESLSLTFHGHDLIVDSDLEXXXXXXXXXXXXXXCGKST 444
K + R TGVLASHP S D+HI +LSLTFHG +L+ D+ LE GKS
Sbjct: 72 KAAARAVTGVLASHPNSTDVHIINLSLTFHGQELLSDTKLELNSGRRYGLIGLNGIGKSM 131
Query: 445 LLTAIGCRELPIPEHMDIYHLSHEIEASDMSALQAIVSCDEERVKLEKEAEILAAQDDGG 624
LL+AIG RE+PIPEH+DIYHL+ E+ S+ + LQ ++ D ER LE+EAE LA +D
Sbjct: 132 LLSAIGKREVPIPEHIDIYHLTREMPPSEKTPLQCVMEVDTERAMLEREAERLA-HEDAE 190
Query: 625 GEALERVYERLDAMDAGTAEKRAAEILFGLGFDKQMQAKKTKDFSGGWRMRIALARALFM 804
E L +YERL+ +DA AE RA+ IL GLGF MQ KK KDFSGGWRMR+ALARALF+
Sbjct: 191 CEKLMELYERLEELDADKAEMRASRILHGLGFTPAMQRKKLKDFSGGWRMRVALARALFI 250
Query: 805 NPTILLLDEPTNHLDLEACVWLEETLKKFDRILVVISHSQDFLNGVCTNIIHMQNKKLKL 984
P +LLLDEPTNHLDL+ACVWLEE LK F RILV++SHSQDFLNGVCTNIIHM NKKLK
Sbjct: 251 RPFMLLLDEPTNHLDLDACVWLEEELKTFKRILVLVSHSQDFLNGVCTNIIHMHNKKLKY 310
Query: 985 YTGNYDQYVQTRSDLEENQMKQYKWEQEQIASMKEYIARFGHGSAKLARQAQSKEKTLAK 1164
YTGNYDQYV+TR +LEENQMK++ WEQ+QIA MK YIARFGHGSAKLARQAQSKEKTL K
Sbjct: 311 YTGNYDQYVKTRLELEENQMKRFHWEQDQIAHMKNYIARFGHGSAKLARQAQSKEKTLQK 370
Query: 1165 MERGGLTEKVVRDRVLVFRFTDVGKLPPPVLQFVEVKFGYTPDN-LIYKSLDFGVDLDSR 1341
M GLTE+VV D+ L F F GK+PPPV+ V F YT D IY +L+FG+DLD+R
Sbjct: 371 MMASGLTERVVSDKTLSFYFPPCGKIPPPVIMVQNVSFKYTKDGPCIYNNLEFGIDLDTR 430
Query: 1342 IALVGPNGAGKSTLLKLMTGDLAPLDGMVRRHNHLRIAQYHQHLADKLDLDMPALAYMMK 1521
+ALVGPNGAGKSTLLKL+TG+L P DGM+R+H+H++I +YHQHL ++LDLD+ L YMMK
Sbjct: 431 VALVGPNGAGKSTLLKLLTGELLPTDGMIRKHSHVKIGRYHQHLQEQLDLDLSPLEYMMK 490
Query: 1522 EYPG-TEEEKMRAAVGRFGLSGKAQVMPMRNLSDGQRSRVIFAWLAYRQPQLLLLDEPTN 1698
YP E+E+MR +GR+GL+GK QV P+RNLSDGQ+ RV AWLA++ P +L LDEPTN
Sbjct: 491 CYPEIKEKEEMRKIIGRYGLTGKQQVSPIRNLSDGQKCRVCLAWLAWQNPHMLFLDEPTN 550
Query: 1699 HLDIETIDSLAEALNEWDGGLVLVSHDFRLINQVAQEIWVCENQAVTRWEGDIMDFKAHL 1878
HLDIETID+LA+A+NE++GG++LVSHDFRLI QVAQEIWVCE Q +T+W GDI+ +K HL
Sbjct: 551 HLDIETIDALADAINEFEGGMMLVSHDFRLIQQVAQEIWVCEKQTITKWPGDILAYKEHL 610
Query: 1879 KSK 1887
KSK
Sbjct: 611 KSK 613
>sptr|Q802V8|Q802V8 Similar to ATP-binding cassette, sub-family F
(GCN20), member 2.
Length = 613
Score = 708 bits (1828), Expect = 0.0
Identities = 349/543 (64%), Positives = 424/543 (78%), Gaps = 2/543 (0%)
Frame = +1
Query: 265 KLSDRTCTGVLASHPLSRDIHIESLSLTFHGHDLIVDSDLEXXXXXXXXXXXXXXCGKST 444
K R TGVLASHP S D+HI SLSLTFHG +L+ D+ LE GKS
Sbjct: 64 KTEARAVTGVLASHPNSTDVHISSLSLTFHGQELLSDTSLELNSGRRYGLIGLNGTGKSM 123
Query: 445 LLTAIGCRELPIPEHMDIYHLSHEIEASDMSALQAIVSCDEERVKLEKEAEILAAQDDGG 624
LL+AI RE+PIPEH+DIYHL+ E+ S+ +ALQ ++ DEER+KLEKEAE LA +D
Sbjct: 124 LLSAISHREVPIPEHIDIYHLTREMAPSEKTALQCVMEVDEERIKLEKEAERLA-HEDSE 182
Query: 625 GEALERVYERLDAMDAGTAEKRAAEILFGLGFDKQMQAKKTKDFSGGWRMRIALARALFM 804
E L +YERL+ +DA A RA+ IL GLGF MQ KK KDFSGGWRMR+ALARALF+
Sbjct: 183 CEKLMELYERLEELDADKASMRASRILHGLGFTTSMQQKKLKDFSGGWRMRVALARALFL 242
Query: 805 NPTILLLDEPTNHLDLEACVWLEETLKKFDRILVVISHSQDFLNGVCTNIIHMQNKKLKL 984
P +LLLDEPTNHLDL+ACVWLEE L +F RILV+ISHSQDFLNGVCTNIIH+ +KLK
Sbjct: 243 KPFMLLLDEPTNHLDLDACVWLEEELSQFKRILVLISHSQDFLNGVCTNIIHLHQRKLKY 302
Query: 985 YTGNYDQYVQTRSDLEENQMKQYKWEQEQIASMKEYIARFGHGSAKLARQAQSKEKTLAK 1164
YTGNYDQYV+TR +LEENQMK++ WEQ+QIA MK YIARFGHGSAKLARQAQSKEKTL K
Sbjct: 303 YTGNYDQYVKTREELEENQMKRFNWEQDQIAHMKNYIARFGHGSAKLARQAQSKEKTLQK 362
Query: 1165 MERGGLTEKVVRDRVLVFRFTDVGKLPPPVLQFVEVKFGYTPDN-LIYKSLDFGVDLDSR 1341
M GLTE+VV D+ L F F GK+PPPV+ V F Y+ + IYK+L+FG+DLD+R
Sbjct: 363 MVASGLTERVVNDKTLSFYFPPCGKIPPPVIMVQNVSFRYSENTPYIYKNLEFGIDLDTR 422
Query: 1342 IALVGPNGAGKSTLLKLMTGDLAPLDGMVRRHNHLRIAQYHQHLADKLDLDMPALAYMMK 1521
+ALVGPNGAGKSTLLKL+TG+L P DGM+R+H+H++I +YHQHL ++L+LD+ L YMMK
Sbjct: 423 VALVGPNGAGKSTLLKLLTGELLPSDGMIRKHSHVKIGRYHQHLTEQLELDLSPLEYMMK 482
Query: 1522 EYPG-TEEEKMRAAVGRFGLSGKAQVMPMRNLSDGQRSRVIFAWLAYRQPQLLLLDEPTN 1698
YP E+E+MR +GR+GL+GK QV P+RNLSDGQ+ RV FAWLA + P +L LDEPTN
Sbjct: 483 CYPEIKEKEEMRKIIGRYGLTGKQQVSPIRNLSDGQKCRVCFAWLARQNPHMLFLDEPTN 542
Query: 1699 HLDIETIDSLAEALNEWDGGLVLVSHDFRLINQVAQEIWVCENQAVTRWEGDIMDFKAHL 1878
HLDIETID+LAEA+N+++GG++LVSHDFRL QVAQEIWVCE Q +T+W DI+ +K HL
Sbjct: 543 HLDIETIDALAEAINDFEGGMMLVSHDFRLTQQVAQEIWVCEKQTITKWSRDILAYKEHL 602
Query: 1879 KSK 1887
KSK
Sbjct: 603 KSK 605
>sptr|Q9VXR5|Q9VXR5 CG9281 protein (LD02975P).
Length = 611
Score = 700 bits (1806), Expect = 0.0
Identities = 347/540 (64%), Positives = 425/540 (78%), Gaps = 3/540 (0%)
Frame = +1
Query: 277 RTCTGVLASHPLSRDIHIESLSLTFHGHDLIVDSDLEXXXXXXXXXXXXXXCGKSTLLTA 456
R+CTG LA HP SRD+ I + S+TF G +L+ D+ LE CGKS+LL
Sbjct: 59 RSCTGSLAVHPRSRDVKIANFSITFFGSELLQDTMLELNCGRRYGLIGLNGCGKSSLLAV 118
Query: 457 IGCRELPIPEHMDIYHLSHEIEASDMSALQAIVSCDEERVKLEKEAEILA-AQDDGGGEA 633
+G RE+P+P H+DI+HL+ EI AS SALQ ++ DEER+KLEK AE LA +++D E
Sbjct: 119 LGGREVPVPPHIDIFHLTREIPASSKSALQCVMEVDEERIKLEKLAEELAMSEEDDAQEQ 178
Query: 634 LERVYERLDAMDAGTAEKRAAEILFGLGFDKQMQAKKTKDFSGGWRMRIALARALFMNPT 813
L +YERLD M A AE +AA IL GLGFDK MQ K+ KDFSGGWRMRIALARALF+ P
Sbjct: 179 LIDIYERLDDMSADLAEVKAARILHGLGFDKAMQQKQAKDFSGGWRMRIALARALFVKPH 238
Query: 814 ILLLDEPTNHLDLEACVWLEETLKKFDRILVVISHSQDFLNGVCTNIIHMQNKKLKLYTG 993
+LLLDEPTNHLDL+ACVWLEE LK + RILV+ISHSQDFLNGVCTNIIH+ K+LK YTG
Sbjct: 239 LLLLDEPTNHLDLDACVWLEEELKTYKRILVLISHSQDFLNGVCTNIIHLTGKRLKYYTG 298
Query: 994 NYDQYVQTRSDLEENQMKQYKWEQEQIASMKEYIARFGHGSAKLARQAQSKEKTLAKMER 1173
NY+ +V+TR +L ENQMKQY WEQ+QI+ MK YIARFGHGSAKLARQAQSKEKTLAKM
Sbjct: 299 NYEAFVRTRMELLENQMKQYNWEQDQISHMKNYIARFGHGSAKLARQAQSKEKTLAKMVA 358
Query: 1174 GGLTEKVVRDRVLVFRFTDVGKLPPPVLQFVEVKFGYTPDN-LIYKSLDFGVDLDSRIAL 1350
GLTEKV D+VL F F GK+PPPV+ V F Y + IYK+L+FG+DLD+R+AL
Sbjct: 359 QGLTEKVSDDKVLNFYFPSCGKVPPPVIMVQNVNFRYNDETPWIYKNLEFGIDLDTRLAL 418
Query: 1351 VGPNGAGKSTLLKLMTGDLAPLDGMVRRHNHLRIAQYHQHLADKLDLDMPALAYMMKEYP 1530
VGPNGAGKSTLLKL+ GDL P GM+R+++HLRIA+YHQHL + LDLD L YMM+ +P
Sbjct: 419 VGPNGAGKSTLLKLLYGDLVPTSGMIRKNSHLRIARYHQHLHELLDLDASPLEYMMRAFP 478
Query: 1531 GT-EEEKMRAAVGRFGLSGKAQVMPMRNLSDGQRSRVIFAWLAYRQPQLLLLDEPTNHLD 1707
E+E+MR +GR+GL+G+ QV P+R LSDGQR RV+FAWLA++ P LLLLDEPTNHLD
Sbjct: 479 DVKEKEEMRKIIGRYGLTGRQQVCPIRQLSDGQRCRVVFAWLAWQVPHLLLLDEPTNHLD 538
Query: 1708 IETIDSLAEALNEWDGGLVLVSHDFRLINQVAQEIWVCENQAVTRWEGDIMDFKAHLKSK 1887
+ETID+LA+A+N++DGG+VLVSHDFRLINQVA+EIWVCE + VT+W+G I+D+K HLK+K
Sbjct: 539 METIDALADAINDFDGGMVLVSHDFRLINQVAEEIWVCEKETVTKWKGGILDYKDHLKNK 598
>sptr|Q7PQX1|Q7PQX1 ENSANGP00000010790 (Fragment).
Length = 620
Score = 690 bits (1781), Expect = 0.0
Identities = 343/538 (63%), Positives = 416/538 (77%), Gaps = 3/538 (0%)
Frame = +1
Query: 277 RTCTGVLASHPLSRDIHIESLSLTFHGHDLIVDSDLEXXXXXXXXXXXXXXCGKSTLLTA 456
R CTG LA HP SRDI + S+TF G +++ D+ LE CGKS+LL+
Sbjct: 65 RACTGSLAVHPRSRDIKFSNFSITFFGSEMLQDTMLELNCGRRYGLLGANGCGKSSLLSV 124
Query: 457 IGCRELPIPEHMDIYHLSHEIEASDMSALQAIVSCDEERVKLEKEAEILAAQ-DDGGGEA 633
+G RE+PIP+H+DI+HL+ EI AS SA+Q ++ DEER+KLEK A+ L Q DD E
Sbjct: 125 LGNREVPIPDHIDIFHLTREIPASSKSAIQCVMEVDEERIKLEKMADALVEQEDDEAQEQ 184
Query: 634 LERVYERLDAMDAGTAEKRAAEILFGLGFDKQMQAKKTKDFSGGWRMRIALARALFMNPT 813
L +Y+RLD M A AE +A+ IL GLGFDK MQ K KDFSGGWRMRIALARALF+ P
Sbjct: 185 LMDIYDRLDEMSADQAEAKASRILHGLGFDKDMQQKAAKDFSGGWRMRIALARALFVKPH 244
Query: 814 ILLLDEPTNHLDLEACVWLEETLKKFDRILVVISHSQDFLNGVCTNIIHMQNKKLKLYTG 993
+LLLDEPTNHLDL+ACVWLEE LK + RILV+ISHSQDFLNGVCTNIIHM K+LK YTG
Sbjct: 245 LLLLDEPTNHLDLDACVWLEEELKTYKRILVIISHSQDFLNGVCTNIIHMTQKRLKYYTG 304
Query: 994 NYDQYVQTRSDLEENQMKQYKWEQEQIASMKEYIARFGHGSAKLARQAQSKEKTLAKMER 1173
NY+Q+V+TR +L ENQMKQY WEQ+QIA MK YIARFGHGSAKLARQAQSKEKTLAKM
Sbjct: 305 NYEQFVKTRLELLENQMKQYNWEQDQIAHMKNYIARFGHGSAKLARQAQSKEKTLAKMVA 364
Query: 1174 GGLTEKVVRDRVLVFRFTDVGKLPPPVLQFVEVKFGYTPDN-LIYKSLDFGVDLDSRIAL 1350
GLTEK V ++ L F F G +PPPV+ V F Y IYK+L+FG+DLD+R+AL
Sbjct: 365 QGLTEKAVDEKQLNFCFPSCGTIPPPVIMVQNVSFRYNDKTPYIYKNLEFGIDLDTRLAL 424
Query: 1351 VGPNGAGKSTLLKLMTGDLAPLDGMVRRHNHLRIAQYHQHLADKLDLDMPALAYMMKEYP 1530
VGPNGAGKSTLLKL+ GDL P GM+R+++HLRIA+YHQHL + LD+D L YM+K +P
Sbjct: 425 VGPNGAGKSTLLKLLYGDLVPTSGMIRKNSHLRIARYHQHLHELLDMDASPLDYMLKNFP 484
Query: 1531 G-TEEEKMRAAVGRFGLSGKAQVMPMRNLSDGQRSRVIFAWLAYRQPQLLLLDEPTNHLD 1707
E E+MR VGR+GL+G+ QV P+R LSDGQR RV+FA+LA+++P LLLLDEPTNHLD
Sbjct: 485 EVVEREEMRKIVGRYGLTGRQQVCPIRQLSDGQRCRVVFAYLAWKKPHLLLLDEPTNHLD 544
Query: 1708 IETIDSLAEALNEWDGGLVLVSHDFRLINQVAQEIWVCENQAVTRWEGDIMDFKAHLK 1881
+ETID+LAEA+N+++GGLVLVSHDFRLINQVA+EIWVCE VT+W G I+D+K HLK
Sbjct: 545 METIDALAEAINDFEGGLVLVSHDFRLINQVAEEIWVCEKGTVTKWNGGILDYKEHLK 602
>sptr|Q9XTD9|Q9XTD9 T27E9.7 protein.
Length = 622
Score = 660 bits (1704), Expect = 0.0
Identities = 325/540 (60%), Positives = 413/540 (76%), Gaps = 3/540 (0%)
Frame = +1
Query: 277 RTCTGVLASHPLSRDIHIESLSLTFHGHDLIVDSDLEXXXXXXXXXXXXXXCGKSTLLTA 456
R+ G L S P D +ESL++TFHG +++VD+ LE GKST++ A
Sbjct: 67 RSVAGSLTSDPKGLDHRVESLTITFHGREIVVDTKLELNRGRRYGLIGLNGSGKSTIIQA 126
Query: 457 IGCRELPIPEHMDIYHLSHEIEASDMSALQAIVSCDEERVKLEKEAEILAAQ-DDGGGEA 633
I +E+PIPE +D+Y +S E+ AS+ +ALQA+V D R +LE AE LA+Q D+ +
Sbjct: 127 IYNKEMPIPESVDMYLVSREMPASEKTALQAVVDVDSVRKELEHLAEQLASQTDEESQDK 186
Query: 634 LERVYERLDAMDAGTAEKRAAEILFGLGFDKQMQAKKTKDFSGGWRMRIALARALFMNPT 813
L VYERLD MDA AEK+AAEIL GLGF K MQ KK KDFSGGWRMRIALARALF+ P+
Sbjct: 187 LMDVYERLDEMDASLAEKKAAEILHGLGFTKTMQMKKCKDFSGGWRMRIALARALFLKPS 246
Query: 814 ILLLDEPTNHLDLEACVWLEETLKKFDRILVVISHSQDFLNGVCTNIIHMQNKKLKLYTG 993
+LLLDEPTNHLDLEACVWLEE L ++ R L+V+SHSQDF+NGVCTNIIH+ K+L Y G
Sbjct: 247 VLLLDEPTNHLDLEACVWLEEELAQYKRTLLVVSHSQDFMNGVCTNIIHLFQKQLVYYGG 306
Query: 994 NYDQYVQTRSDLEENQMKQYKWEQEQIASMKEYIARFGHGSAKLARQAQSKEKTLAKMER 1173
NYDQ+V+TR +L ENQ K+Y WEQ Q+ MK+Y+ARFGHGSAKLARQAQSKEKT+AKM
Sbjct: 307 NYDQFVKTRLELLENQQKRYNWEQSQLQHMKDYVARFGHGSAKLARQAQSKEKTMAKMIA 366
Query: 1174 GGLTEKVVRDRVLVFRFTDVGKLPPPVLQFVEVKFGYTPDN-LIYKSLDFGVDLDSRIAL 1350
GGL EK V + V F F D G++PPPV+ V F Y + IYK +DFG+DLD+RIAL
Sbjct: 367 GGLAEKAVTETVKQFYFFDAGEIPPPVIMVQHVSFRYNENTPWIYKDIDFGIDLDTRIAL 426
Query: 1351 VGPNGAGKSTLLKLMTGDLAPLDGMVRRHNHLRIAQYHQHLADKLDLDMPALAYMMKEYP 1530
VGPNGAGKSTLLKL+ D+ P DG++RRH+H +I +YHQHL ++L LD+ AL +MMKE+P
Sbjct: 427 VGPNGAGKSTLLKLLCTDVMPTDGLIRRHSHCKIGRYHQHLHEELPLDLSALEFMMKEFP 486
Query: 1531 GT-EEEKMRAAVGRFGLSGKAQVMPMRNLSDGQRSRVIFAWLAYRQPQLLLLDEPTNHLD 1707
E+E+MR VGR+G++G+ QV PM+ LSDGQR RV FAWLA++QP LLLLDEPTNHLD
Sbjct: 487 DVKEKEEMRKIVGRYGITGREQVCPMKQLSDGQRCRVSFAWLAWQQPHLLLLDEPTNHLD 546
Query: 1708 IETIDSLAEALNEWDGGLVLVSHDFRLINQVAQEIWVCENQAVTRWEGDIMDFKAHLKSK 1887
+E+ID+LAEA+N + GG++LVSHDFRL++QVA+E+WVC+NQ + +W+GDI FK HL+ +
Sbjct: 547 MESIDALAEAINCFPGGMILVSHDFRLVSQVAEEVWVCDNQGILKWDGDIFSFKEHLRKQ 606
>sw|P40024|YEM6_YEAST Probable ATP-dependent transporter YER036C.
Length = 610
Score = 570 bits (1469), Expect = e-161
Identities = 297/542 (54%), Positives = 382/542 (70%), Gaps = 5/542 (0%)
Frame = +1
Query: 268 LSDRTCTGVLASHPLSRDIHIESLSLTFHGHDLIVDSDLEXXXXXXXXXXXXXXCGKSTL 447
LSDR TGVL+S SRDI + S+SL FHG LI DS LE CGKST
Sbjct: 64 LSDRVVTGVLSSLETSRDIKLSSVSLLFHGKVLIQDSGLELNYGRRYGLLGENGCGKSTF 123
Query: 448 LTAIGCRELPIPEHMDIYHLSHEIEASDMSALQAIVS-CDEERVKLEKEAEILAAQDDGG 624
L A+ RE PIPEH+DIY L E S++SAL +V+ E ++E E +D
Sbjct: 124 LKALATREYPIPEHIDIYLLDEPAEPSELSALDYVVTEAQHELKRIEDLVEKTILEDGPE 183
Query: 625 GEALERVYERLDAMDAGTAEKRAAEILFGLGFDKQMQAKKTKDFSGGWRMRIALARALFM 804
E LE +YER+D++D T E RAA IL GLGF+K+ KKTKD SGGW+MR+ALA+ALF+
Sbjct: 184 SELLEPLYERMDSLDPDTFESRAAIILIGLGFNKKTILKKTKDMSGGWKMRVALAKALFV 243
Query: 805 NPTILLLDEPTNHLDLEACVWLEETLKKFDRILVVISHSQDFLNGVCTNIIHMQNKKLKL 984
PT+LLLD+PT HLDLEACVWLEE LK+FDR LV++SHSQDFLNGVCTN+I M+ +KL
Sbjct: 244 KPTLLLLDDPTAHLDLEACVWLEEYLKRFDRTLVLVSHSQDFLNGVCTNMIDMRAQKLTA 303
Query: 985 YTGNYDQYVQTRSDLEENQMKQYKWEQEQIASMKEYIARFGHGSAKLARQAQSKEKTLAK 1164
Y GNYD Y +TRS+LE NQMKQY +QE+I +K++IA G A L +QA+S++K L K
Sbjct: 304 YGGNYDSYHKTRSELETNQMKQYNKQQEEIQHIKKFIASAG-TYANLVKQAKSRQKILDK 362
Query: 1165 MERGGLTEKVVRDRVLVFRFTDVGKLPPPVLQFVEVKFGY--TPDNLIYKSLDFGVDLDS 1338
ME GL + VV D+V FRF V +LPPPVL F ++ F Y P +Y+ L+FGVD+DS
Sbjct: 363 MEADGLVQPVVPDKVFSFRFPQVERLPPPVLAFDDISFHYESNPSENLYEHLNFGVDMDS 422
Query: 1339 RIALVGPNGAGKSTLLKLMTGDLAPLDGMVRRHNHLRIAQYHQHLADKLDLDMPALAYMM 1518
RIALVGPNG GKSTLLK+MTG+L P G V RH H+++ Y QH D+LDL AL ++
Sbjct: 423 RIALVGPNGVGKSTLLKIMTGELTPQSGRVSRHTHVKLGVYSQHSQDQLDLTKSALEFVR 482
Query: 1519 KEYPGTEEEKM--RAAVGRFGLSGKAQVMPMRNLSDGQRSRVIFAWLAYRQPQLLLLDEP 1692
+Y ++ R +GR+GL+G+ Q + M LS+GQRSRV+FA LA QP +LLLDEP
Sbjct: 483 DKYSNISQDFQFWRGQLGRYGLTGEGQTVQMATLSEGQRSRVVFALLALEQPNVLLLDEP 542
Query: 1693 TNHLDIETIDSLAEALNEWDGGLVLVSHDFRLINQVAQEIWVCENQAVTRWEGDIMDFKA 1872
TN LDI TIDSLA+A+NE++GG+V+VSHDFRL++++AQ+I+V EN+ TRW+G I+ +K
Sbjct: 543 TNGLDIPTIDSLADAINEFNGGVVVVSHDFRLLDKIAQDIFVVENKTATRWDGSILQYKN 602
Query: 1873 HL 1878
L
Sbjct: 603 KL 604
>sptr|Q8T6B7|Q8T6B7 Non-transporter ABC protein AbcF2.
Length = 593
Score = 567 bits (1460), Expect = e-160
Identities = 286/547 (52%), Positives = 381/547 (69%), Gaps = 5/547 (0%)
Frame = +1
Query: 262 LKLSDRTCTGVLASHPLSRDIHIESLSLTFHGHDLIVDSDLEXXXXXXXXXXXXXXCGKS 441
++L+ T TG LAS SRD+ IE ++LTFHG +L+ D+ +E CGKS
Sbjct: 33 MRLNAFTATGALASKESSRDVKIEQVTLTFHGKELLSDTTVEINFGRRYGLIGQNGCGKS 92
Query: 442 TLLTAIGCRELPIPEHMDIYHLSHEIEASDMSALQAIVSCDEERVK-LEKEAEILAAQDD 618
T + RELPIPEH+DI+HLS E S+ +ALQ+++ E+ VK LE E L +
Sbjct: 93 TFFQCLAVRELPIPEHIDIFHLSEEAHPSERTALQSVIDDAEKEVKRLEVLEERLLEEQG 152
Query: 619 GGGEALERVYERLDAMDAGTAEKRAAEILFGLGFDKQMQAKKTKDFSGGWRMRIALARAL 798
E L VYERL+ +D T RA+EIL GLGF Q KKTKD SGGWRMR++LA+AL
Sbjct: 153 PESEELFDVYERLENLDPTTFVPRASEILIGLGFTSQTMLKKTKDLSGGWRMRVSLAKAL 212
Query: 799 FMNPTILLLDEPTNHLDLEACVWLEETLKKFDRILVVISHSQDFLNGVCTNIIHMQNKKL 978
F+ PT+LLLDEPTNHLDL ACVWLE+ L +DR L++ISHSQDFLN VCTNIIHM KL
Sbjct: 213 FIKPTLLLLDEPTNHLDLGACVWLEDYLANYDRSLIIISHSQDFLNAVCTNIIHMTQSKL 272
Query: 979 KLYTGNYDQYVQTRSDLEENQMKQYKWEQEQIASMKEYIARFGHGSAKLARQAQSKEKTL 1158
K Y GNYD +V+T+++LE NQMK Y +QE+IA +K +IA G S L RQ +SK+K +
Sbjct: 273 KYYGGNYDNFVKTKAELEVNQMKAYHKQQEEIAHIKSFIASCGTYS-NLVRQGKSKQKII 331
Query: 1159 AKMERGGLTEKVVRDRVLVFRFTDVGKLPPPVLQFVEVKFGYT--PDNLIYKSLDFGVDL 1332
KME GL E+V D++ F F G+L PP++ F V F Y+ +++Y++LD +DL
Sbjct: 332 DKMEEAGLVERVQEDKIFNFSFPPCGELAPPIMHFDNVTFSYSGKEADVLYRNLDLAIDL 391
Query: 1333 DSRIALVGPNGAGKSTLLKLMTGDLAPLDGMVRRHNHLRIAQYHQHLADKLDLDMPALAY 1512
DSRIALVGPNGAGKSTLLKLM G ++P G +++H+HL++A+YHQH + LDL L +
Sbjct: 392 DSRIALVGPNGAGKSTLLKLMVGQISPTQGFIKKHSHLKMARYHQHAHEVLDLTATPLDF 451
Query: 1513 MMKEYP--GTEEEKMRAAVGRFGLSGKAQVMPMRNLSDGQRSRVIFAWLAYRQPQLLLLD 1686
+ ++ + E+ R +GRFG++GKAQ + +SDG +SR+IF +A P LLLLD
Sbjct: 452 VRSKFAHMNKDTEEWRREIGRFGVTGKAQTEAIGCMSDGIKSRLIFCLMALENPHLLLLD 511
Query: 1687 EPTNHLDIETIDSLAEALNEWDGGLVLVSHDFRLINQVAQEIWVCENQAVTRWEGDIMDF 1866
EPTNHLD+E IDSLA A+N + GG++LVSHDFRLI+QVA+EIWVC+N+ +T+W GDI +
Sbjct: 512 EPTNHLDMECIDSLALAINSFPGGMILVSHDFRLISQVAKEIWVCDNKTITKWAGDITSY 571
Query: 1867 KAHLKSK 1887
K HLK++
Sbjct: 572 KNHLKAQ 578
>sptr|Q872I6|Q872I6 Probable iron inhibited ABC transporter 2
(Hypothetical protein).
Length = 613
Score = 559 bits (1441), Expect = e-158
Identities = 290/545 (53%), Positives = 383/545 (70%), Gaps = 5/545 (0%)
Frame = +1
Query: 268 LSDRTCTGVLASHPLSRDIHIESLSLTFHGHDLIVDSDLEXXXXXXXXXXXXXXCGKSTL 447
LSDR TGVLAS S+D+ I S+SL FHG LI D+ LE CGKSTL
Sbjct: 63 LSDRVTTGVLASTKASKDVKITSVSLVFHGRVLIKDATLELTLGRRYGLLGENGCGKSTL 122
Query: 448 LTAIGCRELPIPEHMDIYHLSHEIEASDMSALQAIVS-CDEERVKLEKEAEILAAQDDGG 624
L AI RE PIPEH+DIY L+ S++ AL +V+ E +LE AE + ++
Sbjct: 123 LKAIAAREYPIPEHVDIYLLNEGAPPSELGALDWVVTEAKNELARLESVAEKILEEEGPD 182
Query: 625 GEALERVYERLDAMDAGTAEKRAAEILFGLGFDKQMQAKKTKDFSGGWRMRIALARALFM 804
LE +Y+ +D MD T E RA+ IL GLGF+KQ KKTKD SGGWRMR+ALA+ALF+
Sbjct: 183 SPLLEDLYDHIDKMDPSTFETRASLILTGLGFNKQTIQKKTKDMSGGWRMRVALAKALFV 242
Query: 805 NPTILLLDEPTNHLDLEACVWLEETLKKFDRILVVISHSQDFLNGVCTNIIHMQNKKLKL 984
PT+LLLD+PT HLDLEACVWLEE LKK+DR LV++SHS+DFLNGVCT++I M+ + L
Sbjct: 243 KPTLLLLDDPTAHLDLEACVWLEEYLKKWDRTLVLVSHSEDFLNGVCTSMIDMRLQTLIY 302
Query: 985 YTGNYDQYVQTRSDLEENQMKQYKWEQEQIASMKEYIARFGHGSAKLARQAQSKEKTLAK 1164
Y GNYD Y +TR++ E NQMK Y+ +Q++IA +K++IA G A L RQA+S++K L K
Sbjct: 303 YGGNYDTYHKTRAEQETNQMKAYQKQQDEIAHIKKFIASAG-TYANLVRQAKSRQKILDK 361
Query: 1165 MERGGLTEKVVRDRVLVFRFTDVGKLPPPVLQFVEVKFGYT--PDNLIYKSLDFGVDLDS 1338
ME G + V +DRV FRF DV KLPPPVL F V F Y+ P + +Y+ +D G D+DS
Sbjct: 362 MEADGFIQPVKQDRVFSFRFADVEKLPPPVLSFDNVTFSYSGDPKDDLYRHIDLGFDMDS 421
Query: 1339 RIALVGPNGAGKSTLLKLMTGDLAPLDGMVRRHNHLRIAQYHQHLADKLDLDMPALAYMM 1518
R ALVGPNG GKSTLL+LMTG L+P DG+V+RH HL++ Y QH A++LDL AL ++
Sbjct: 422 RTALVGPNGVGKSTLLRLMTGKLSPTDGVVQRHTHLKLGLYSQHSAEQLDLTKSALDFVR 481
Query: 1519 KEYPGTEEEKM--RAAVGRFGLSGKAQVMPMRNLSDGQRSRVIFAWLAYRQPQLLLLDEP 1692
+YP ++ R +G++GLSG+AQ + LSDGQ+SR++FA LA P +LLLDEP
Sbjct: 482 DKYPDKSQDYQYWRQQLGKYGLSGEAQTSLIGTLSDGQKSRIVFALLAIESPNMLLLDEP 541
Query: 1693 TNHLDIETIDSLAEALNEWDGGLVLVSHDFRLINQVAQEIWVCENQAVTRWEGDIMDFKA 1872
TN LDI TIDSLA+A+N + GG+++VSHDFRL++++A++I VCEN+ + +WEG I D+K
Sbjct: 542 TNGLDIPTIDSLADAINAFTGGVIVVSHDFRLLDKIAKQILVCENKTIKQWEGSISDYKN 601
Query: 1873 HLKSK 1887
+L+ K
Sbjct: 602 YLRKK 606
>sptr|Q17340|Q17340 Probable ATP-dependent transporter GNC20-2
(Fragment).
Length = 535
Score = 554 bits (1427), Expect = e-156
Identities = 280/474 (59%), Positives = 352/474 (74%), Gaps = 3/474 (0%)
Frame = +1
Query: 265 KLSDRTCTGVLASHPLSRDIHIESLSLTFHGHDLIVDSDLEXXXXXXXXXXXXXXCGKST 444
K + R+ G L S P D +ESL++TFHG +++VD+ LE GKST
Sbjct: 62 KRAARSVAGSLTSDPKGLDHRVESLTITFHGREIVVDTKLELNRGRRYGLIGLNGSGKST 121
Query: 445 LLTAIGCRELPIPEHMDIYHLSHEIEASDMSALQAIVSCDEERVKLEKEAEILAAQ-DDG 621
++ AI +E+PIPE +D+Y +S E+ AS+ +ALQA+V D R +LE AE LA+Q D+
Sbjct: 122 IIQAIYNKEMPIPESVDMYLVSREMPASEKTALQAVVDVDSVRKELEHLAEQLASQTDEE 181
Query: 622 GGEALERVYERLDAMDAGTAEKRAAEILFGLGFDKQMQAKKTKDFSGGWRMRIALARALF 801
+ L VYERLD MDA AEK+AAEIL GLGF K MQ KK KDFSGGWRMRI LARALF
Sbjct: 182 SQDKLMDVYERLDEMDASLAEKKAAEILHGLGFTKTMQMKKCKDFSGGWRMRIRLARALF 241
Query: 802 MNPTILLLDEPTNHLDLEACVWLEETLKKFDRILVVISHSQDFLNGVCTNIIHMQNKKLK 981
+ P++LLLDEPTNHLDLEACVWLEE L ++ R L+V+SHSQDF+NGVCTNIIH+ K+L
Sbjct: 242 LKPSVLLLDEPTNHLDLEACVWLEEELAQYKRTLLVVSHSQDFMNGVCTNIIHLFQKQLV 301
Query: 982 LYTGNYDQYVQTRSDLEENQMKQYKWEQEQIASMKEYIARFGHGSAKLARQAQSKEKTLA 1161
Y GNYDQ+V+TR +L ENQ K+Y WEQ Q+ MK+Y+ARFGHGSAKLARQAQSKEKT+A
Sbjct: 302 YYGGNYDQFVKTRLELLENQQKRYNWEQSQLQHMKDYVARFGHGSAKLARQAQSKEKTMA 361
Query: 1162 KMERGGLTEKVVRDRVLVFRFTDVGKLPPPVLQFVEVKFGYTPDN-LIYKSLDFGVDLDS 1338
KM GGL EK V + V F F D G++PPPV+ V F Y + IYK +DFG+DLD+
Sbjct: 362 KMIAGGLAEKAVTETVKQFYFFDAGEIPPPVIMVQHVSFRYNENTPWIYKDIDFGIDLDT 421
Query: 1339 RIALVGPNGAGKSTLLKLMTGDLAPLDGMVRRHNHLRIAQYHQHLADKLDLDMPALAYMM 1518
RIALVGPNGAGKSTLLKL+ D+ P DG++RRH+H +I +YHQHL ++L LD+ AL +MM
Sbjct: 422 RIALVGPNGAGKSTLLKLLCTDVMPTDGLIRRHSHCKIGRYHQHLHEELPLDLSALEFMM 481
Query: 1519 KEYPGT-EEEKMRAAVGRFGLSGKAQVMPMRNLSDGQRSRVIFAWLAYRQPQLL 1677
KE+P E+E+MR VGR+G++G+ QV PM+ LSDGQR RV FAWLA++QP LL
Sbjct: 482 KEFPDVKEKEEMRKIVGRYGITGREQVCPMKQLSDGQRCRVSFAWLAWQQPHLL 535
Score = 63.5 bits (153), Expect = 1e-08
Identities = 51/185 (27%), Positives = 83/185 (44%), Gaps = 27/185 (14%)
Frame = +1
Query: 1339 RIALVGPNGAGKSTLLKLMTGDLAPLDGMVRRHNHLR------------------IAQYH 1464
R L+G NG+GKST+++ + P+ V + R + +
Sbjct: 108 RYGLIGLNGSGKSTIIQAIYNKEMPIPESVDMYLVSREMPASEKTALQAVVDVDSVRKEL 167
Query: 1465 QHLADKL--DLDMPALAYMMKEYPGTEE-------EKMRAAVGRFGLSGKAQVMPMRNLS 1617
+HLA++L D + +M Y +E +K + G + Q+ ++ S
Sbjct: 168 EHLAEQLASQTDEESQDKLMDVYERLDEMDASLAEKKAAEILHGLGFTKTMQMKKCKDFS 227
Query: 1618 DGQRSRVIFAWLAYRQPQLLLLDEPTNHLDIETIDSLAEALNEWDGGLVLVSHDFRLINQ 1797
G R R+ A + +P +LLLDEPTNHLD+E L E L ++ L++VSH +N
Sbjct: 228 GGWRMRIRLARALFLKPSVLLLDEPTNHLDLEACVWLEEELAQYKRTLLVVSHSQDFMNG 287
Query: 1798 VAQEI 1812
V I
Sbjct: 288 VCTNI 292
>sptr|O42943|O42943 Non transporter with ABC binding cassette.
Length = 618
Score = 521 bits (1341), Expect = e-146
Identities = 278/551 (50%), Positives = 374/551 (67%), Gaps = 8/551 (1%)
Frame = +1
Query: 274 DRTCTGVLASHPLSRDIHIESLSLTFHGHDLIVDSDLEXXXXXXXXXXXXXXCGKSTLLT 453
+R+ +GVL S P+SRDI I+S +L+FHG LI ++ +E GKST L
Sbjct: 60 ERSASGVLTSQPMSRDIKIDSYTLSFHGRLLIENATIELNHGQRYGLLGDNGSGKSTFLE 119
Query: 454 AIGCRELPIPEHMDIYHLSHEIEASDMSALQAIVSCDEERV-KLEKEAEILAAQDDGGGE 630
++ R++ PEH+D Y L+ E E SD++A+ I+ +++V KLE E E L+ DD
Sbjct: 120 SVAARDVEYPEHIDSYLLNAEAEPSDVNAVDYIIQSAKDKVQKLEAEIEELSTADDVDDV 179
Query: 631 ALERVYERLDAMDAGTAEKRAAEILFGLGFDKQMQAKKTKDFSGGWRMRIALARALFMNP 810
LE YE LD MD T E +AA IL GLGF ++M AK TKD SGGWRMR+AL+RALF+ P
Sbjct: 180 LLESKYEELDDMDPSTFEAKAAMILHGLGFTQEMMAKPTKDMSGGWRMRVALSRALFIKP 239
Query: 811 TILLLDEPTNHLDLEACVWLEETLKKFDRILVVISHSQDFLNGVCTNIIHMQNKK-LKLY 987
++LLLDEPTNHLDLEA VWLE L K+D+ILVV SHSQDFLN VCTNII + +KK L Y
Sbjct: 240 SLLLLDEPTNHLDLEAVVWLENYLAKYDKILVVTSHSQDFLNNVCTNIIDLTSKKQLVYY 299
Query: 988 TGNYDQYVQTRSDLEENQMKQYKWEQEQIASMKEYIARFGHGSAKLARQAQSKEKTLAKM 1167
GN+D Y++T+ + E NQMK Y +QE+IA +K++IA G A L RQA+SK+K + KM
Sbjct: 300 GGNFDIYMRTKEENETNQMKAYLKQQEEIAHIKKFIASAG-TYANLVRQAKSKQKIIDKM 358
Query: 1168 ERGGLTEKVVRDRVLVFRFTDVGKLPPPVLQFVEVKFGY--TPDNLIYKSLDFGVDLDSR 1341
E GL EK R F F +V KLPPP++ F +V F Y D+ +Y+ L FG+D+DSR
Sbjct: 359 EAAGLVEKPEPPRQFSFEFDEVRKLPPPIIAFNDVAFSYDGNLDHALYRDLSFGIDMDSR 418
Query: 1342 IALVGPNGAGKSTLLKLMTGDLAPLDGMVRRHNHLRIAQYHQHLADKLDLDMPALAYMMK 1521
+A+VG NG GKSTLL L+TG L P++G V R++ L++A+Y QH AD+L D L Y+M
Sbjct: 419 VAIVGKNGTGKSTLLNLITGLLIPIEGNVSRYSGLKMAKYSQHSADQLPYDKSPLEYIMD 478
Query: 1522 EY----PGTEEEKMRAAVGRFGLSGKAQVMPMRNLSDGQRSRVIFAWLAYRQPQLLLLDE 1689
Y P E ++ R+ +G+FGLSG Q +R LSDG +SRV+FA LA QP +LLLDE
Sbjct: 479 TYKPKFPERELQQWRSVLGKFGLSGLHQTSEIRTLSDGLKSRVVFAALALEQPHILLLDE 538
Query: 1690 PTNHLDIETIDSLAEALNEWDGGLVLVSHDFRLINQVAQEIWVCENQAVTRWEGDIMDFK 1869
PTNHLDI +ID+LA+A+N W GG+VLVSHDFRLI QV++E+W +++ V + + I ++K
Sbjct: 539 PTNHLDITSIDALAKAINVWTGGVVLVSHDFRLIGQVSKELWEVKDKKVVKLDCSIEEYK 598
Query: 1870 AHLKSKAGLSD 1902
+ + D
Sbjct: 599 KSMAKEVQSRD 609
Score = 68.6 bits (166), Expect = 4e-10
Identities = 63/258 (24%), Positives = 112/258 (43%), Gaps = 40/258 (15%)
Frame = +1
Query: 1297 LIYKSLDFGVDLDSRIALVGPNGAGKSTLLKLMTG---------------------DLAP 1413
L+ ++ ++ R L+G NG+GKST L+ + D+
Sbjct: 89 LLIENATIELNHGQRYGLLGDNGSGKSTFLESVAARDVEYPEHIDSYLLNAEAEPSDVNA 148
Query: 1414 LDGMVR--RHNHLRIAQYHQHLADKLDLDMPALAYMMKEY----PGTEEEKMRAAVGRFG 1575
+D +++ + ++ + L+ D+D L +E P T E K + G
Sbjct: 149 VDYIIQSAKDKVQKLEAEIEELSTADDVDDVLLESKYEELDDMDPSTFEAKAAMILHGLG 208
Query: 1576 LSGKAQVMPMRNLSDGQRSRVIFAWLAYRQPQLLLLDEPTNHLDIETIDSLAEALNEWDG 1755
+ + P +++S G R RV + + +P LLLLDEPTNHLD+E + L L ++D
Sbjct: 209 FTQEMMAKPTKDMSGGWRMRVALSRALFIKPSLLLLDEPTNHLDLEAVVWLENYLAKYDK 268
Query: 1756 GLVLVSHDFRLINQVAQEI--WVCENQAV-----------TRWEGDIMDFKAHLKSKAGL 1896
LV+ SH +N V I + Q V T+ E + KA+LK + +
Sbjct: 269 ILVVTSHSQDFLNNVCTNIIDLTSKKQLVYYGGNFDIYMRTKEENETNQMKAYLKQQEEI 328
Query: 1897 SD*RAFSSNHELFNSVSR 1950
+ + F ++ + ++ R
Sbjct: 329 AHIKKFIASAGTYANLVR 346
>sptr|Q8T6B8|Q8T6B8 Non-transporter ABC protein AbcF1.
Length = 708
Score = 463 bits (1192), Expect = e-129
Identities = 238/528 (45%), Positives = 352/528 (66%), Gaps = 9/528 (1%)
Frame = +1
Query: 313 SRDIHIESLSLTFHGHDLIVDSDLEXXXXXXXXXXXXXXCGKSTLLTAIGCRELPIPEHM 492
SRDIH+++ ++T+ +DLI++SDL GK+TLL I RE+ I ++
Sbjct: 179 SRDIHVDAFNVTYGKNDLIINSDLNLNYGRKYGLIGRNGTGKTTLLRHIASREIGIDNNL 238
Query: 493 DIYHLSHEIEASDMSALQAIVSCDEERVKLEKEAEILAA----QDDGGGEALERVYERLD 660
I H+ E+ ++ + ++ ++ D ER +L KE + L A + + E L +YE+L+
Sbjct: 239 SILHVEQEVNGNETTVIECVLEADVERDRLLKEEKRLNALPESEKNNLSEKLNSIYEKLN 298
Query: 661 AMDAGTAEKRAAEILFGLGFDKQMQAKKTKDFSGGWRMRIALARALFMNPTILLLDEPTN 840
+DA TAE RAA IL GLGF ++MQ + TK FSGGWRMRI+LARALF+ P +LLLDEPTN
Sbjct: 299 HIDAHTAESRAAAILSGLGFTEEMQQQPTKSFSGGWRMRISLARALFIQPDVLLLDEPTN 358
Query: 841 HLDLEACVWLEETLKKFDRILVVISHSQDFLNGVCTNIIHMQNKKLKLYTGNYDQYVQTR 1020
HLDL AC+WLE L ++R LV++SH ++FLN VCT+I+H+ NKK+ Y GNY + +TR
Sbjct: 359 HLDLFACLWLESYLVNWNRTLVIVSHQREFLNAVCTDIMHLNNKKIDYYKGNYSVFERTR 418
Query: 1021 SDLEENQMKQYKWEQEQIASMKEYIARFGHGSAKLARQAQSKEKTLAKMERGGLTEKVVR 1200
SD + Q + ++ +Q Q ++ +I RF + +AK A+ AQS+ K L +ME G +V+
Sbjct: 419 SDRLKAQQRSFEAQQSQKKHIQAFIDRFRY-NAKRAKMAQSRIKQLERMEDIG---EVLD 474
Query: 1201 DRVLVFRFTDVGKLPPPVLQFVEVKFGYTPDNLIYKSLDFGVDLDSRIALVGPNGAGKST 1380
D + +F + L PP+LQF +V FGYTPD L++K+L+ G+D+ SR+ALVG NGAGK+T
Sbjct: 475 DPTVTLQFLEPEPLAPPILQFQDVSFGYTPDKLLFKNLNLGIDMGSRVALVGANGAGKTT 534
Query: 1381 LLKLMTGDLAPLDGMVRRHNHLRIAQYHQHLADKLDLDMPALAYMMKEYPGTEEEKMRAA 1560
LL+L+ G+L +G+V R+ LR +++ QH D+LDL L + +YPGT + R+
Sbjct: 535 LLRLLCGELEETNGLVIRNGKLRFSRFSQHFVDQLDLSKSPLDNFLAKYPGTTAQTARSH 594
Query: 1561 VGRFGLSGKAQVMPMRNLSDGQRSRVIFAWLAYRQPQLLLLDEPTNHLDIETIDSLAEAL 1740
+G+FGLSG + + LS GQ+SRV+ A ++Y +P +LLLDEP+NHLDI+T+D+L AL
Sbjct: 595 LGKFGLSGDIALRTVNTLSGGQKSRVVLAQISYTKPHILLLDEPSNHLDIDTVDALCHAL 654
Query: 1741 NEWDGGLVLVSHDFRLINQVAQEIWVC-----ENQAVTRWEGDIMDFK 1869
N ++GG++LVSHD RLI+ V EIW E + V ++GD D+K
Sbjct: 655 NVFEGGILLVSHDERLISLVCDEIWYFDGEEGEPKEVKNFDGDWNDYK 702
>sptr|Q9USH9|Q9USH9 Putative ABC transporter.
Length = 822
Score = 446 bits (1148), Expect = e-124
Identities = 237/543 (43%), Positives = 352/543 (64%), Gaps = 7/543 (1%)
Frame = +1
Query: 280 TCTGVLASHPLSRDIHIESLSLTFHGHDLIVDSDLEXXXXXXXXXXXXXXCGKSTLLTAI 459
T TG L S P SRD+ +E LS++ G LI DS+L GKSTLL AI
Sbjct: 262 TVTGNLLSPPNSRDLQVEKLSVSAWGKLLIKDSELNLINGRRYGLIAPNGSGKSTLLHAI 321
Query: 460 GCRELPIPEHMDIYHLSHEIEASDMSALQAIVSCDE-ERVKLEKEAEILAAQDDGGGEAL 636
C +P P +D Y L E ++++ ++A++ +E ER LE E L D L
Sbjct: 322 ACGLIPTPSSLDFYLLDREYIPNELTCVEAVLDINEQERKHLEAMMEDLLDDPDKNAVEL 381
Query: 637 ERVYERLDAMDAGTAEKRAAEILFGLGFDKQMQAKKTKDFSGGWRMRIALARALFMNPTI 816
+ + RL ++ ++ R +IL GL F +M AK+T + SGGWRMRIALAR LF+ PT+
Sbjct: 382 DTIQTRLTDLETENSDHRVYKILRGLQFTDEMIAKRTNELSGGWRMRIALARILFIKPTL 441
Query: 817 LLLDEPTNHLDLEACVWLEETL--KKFDRILVVISHSQDFLNGVCTNIIHMQNKKLKLYT 990
++LDEPTNHLDLEA WLEE L + L++ H+QD LN VCT+IIH+ ++KL Y+
Sbjct: 442 MMLDEPTNHLDLEAVAWLEEYLTHEMEGHTLLITCHTQDTLNEVCTDIIHLYHQKLDYYS 501
Query: 991 GNYDQYVQTRSDLEENQMKQYKWEQEQIASMKEYIARFGHGSAKLAR-QAQSKEKTLAKM 1167
GNYD +++ R++ + K+ + +++ +A ++ + G K A+ + ++ K L K
Sbjct: 502 GNYDTFLKVRAERDVQLAKKARQQEKDMAKLQNKLNMTGSEQQKKAKAKVKAMNKKLEKD 561
Query: 1168 ERGG--LTEKVVRDRVLVFRFTDVGK-LPPPVLQFVEVKFGYTPDNLIYKSLDFGVDLDS 1338
++ G L E++++++ LV RF D G +P P ++F +V F Y I+ L+FG+DL S
Sbjct: 562 KQSGKVLDEEIIQEKQLVIRFEDCGGGIPSPAIKFQDVSFNYPGGPTIFSKLNFGLDLKS 621
Query: 1339 RIALVGPNGAGKSTLLKLMTGDLAPLDGMVRRHNHLRIAQYHQHLADKLDLDMPALAYMM 1518
R+ALVGPNGAGK+TL+KL+ + P G V RH+ LR+A ++QH+ D+LD+ + A+ ++
Sbjct: 622 RVALVGPNGAGKTTLIKLILEKVQPSTGSVVRHHGLRLALFNQHMGDQLDMRLSAVEWLR 681
Query: 1519 KEYPGTEEEKMRAAVGRFGLSGKAQVMPMRNLSDGQRSRVIFAWLAYRQPQLLLLDEPTN 1698
++ E +MR VGR+GL+GK+QV+PM LSDGQR RV+FA+L QP +LLLDEPTN
Sbjct: 682 TKFGNKPEGEMRRIVGRYGLTGKSQVIPMGQLSDGQRRRVLFAFLGMTQPHILLLDEPTN 741
Query: 1699 HLDIETIDSLAEALNEWDGGLVLVSHDFRLINQVAQEIWVCENQAVTRWEGDIMDFKAHL 1878
LDI+TID+LA+ALN +DGG+V ++HDFRLI+QVA+EIW+ +N V ++G+I D+K L
Sbjct: 742 ALDIDTIDALADALNNFDGGVVFITHDFRLIDQVAEEIWIVQNGTVKEFDGEIRDYKMML 801
Query: 1879 KSK 1887
K +
Sbjct: 802 KQQ 804
>sptrnew|AAQ65167|AAQ65167 At1g64550.
Length = 715
Score = 425 bits (1093), Expect = e-117
Identities = 232/545 (42%), Positives = 334/545 (61%), Gaps = 22/545 (4%)
Frame = +1
Query: 316 RDIHIESLSLTFHGHDLIVDSDLEXXXXXXXXXXXXXXCGKSTLLTAIGCRELP-IPEHM 492
RDIH+++ +++ G DLIVD + GK+T L + + IP +
Sbjct: 173 RDIHMDNFNVSVGGRDLIVDGSITLSFGRHYGLVGRNGTGKTTFLRYMAMHAIEGIPTNC 232
Query: 493 DIYHLSHEIEASDMSALQAIVSCDEERVKL-EKEAEILAAQ------------------- 612
I H+ E+ +ALQ +++ D ER KL E+E +ILA Q
Sbjct: 233 QILHVEQEVVGDKTTALQCVLNTDIERTKLLEEEIQILAKQRETEEPTAKDGMPTKDTVE 292
Query: 613 DDGGGEALERVYERLDAMDAGTAEKRAAEILFGLGFDKQMQAKKTKDFSGGWRMRIALAR 792
D + LE +Y+RLDA+DA TAE RAA IL GL F +MQ K T FSGGWRMRIALAR
Sbjct: 293 GDLMSQRLEEIYKRLDAIDAYTAEARAASILAGLSFTPEMQLKATNTFSGGWRMRIALAR 352
Query: 793 ALFMNPTILLLDEPTNHLDLEACVWLEETLKKFDRILVVISHSQDFLNGVCTNIIHMQNK 972
ALF+ P +LLLDEPTNHLDL A +WLE L K+ + +V+SH+++FLN V T+IIH+QN+
Sbjct: 353 ALFIEPDLLLLDEPTNHLDLHAVLWLETYLTKWPKTFIVVSHAREFLNTVVTDIIHLQNQ 412
Query: 973 KLKLYTGNYDQYVQTRSDLEENQMKQYKWEQEQIASMKEYIARFGHGSAKLARQAQSKEK 1152
KL Y GNYD + +TR + +NQ K ++ + + M+ +I +F + +AK A QS+ K
Sbjct: 413 KLSTYKGNYDIFERTREEQVKNQQKAFESSERSRSHMQAFIDKFRY-NAKRASLVQSRIK 471
Query: 1153 TLAKMERGGLTEKVVRDRVLVFRF-TDVGKLPPPVLQFVEVKFGYTPDNLIYKSLDFGVD 1329
L ++ ++V+ D F F T K PP++ F + FGY L++++L+FG+D
Sbjct: 472 ALDRLAH---VDQVINDPDYKFEFPTPDDKPGPPIISFSDASFGYPGGPLLFRNLNFGID 528
Query: 1330 LDSRIALVGPNGAGKSTLLKLMTGDLAPLDGMVRRHNHLRIAQYHQHLADKLDLDMPALA 1509
LDSRIA+VGPNG GKST+LKL++GDL P G V R +R+A + QH D LDL L
Sbjct: 529 LDSRIAMVGPNGIGKSTILKLISGDLQPSSGTVFRSAKVRVAVFSQHHVDGLDLSSNPLL 588
Query: 1510 YMMKEYPGTEEEKMRAAVGRFGLSGKAQVMPMRNLSDGQRSRVIFAWLAYRQPQLLLLDE 1689
YMM+ YPG E+K+R+ +G G++G + PM LS GQ+SRV FA + +++P LLLLDE
Sbjct: 589 YMMRCYPGVPEQKLRSHLGSLGVTGNLALQPMYTLSGGQKSRVAFAKITFKKPHLLLLDE 648
Query: 1690 PTNHLDIETIDSLAEALNEWDGGLVLVSHDFRLINQVAQEIWVCENQAVTRWEGDIMDFK 1869
P+NHLD++ +++L + L + GG+ +VSHD LI+ E+WV + + + G D+K
Sbjct: 649 PSNHLDLDAVEALIQGLVLFQGGICMVSHDEHLISGSVDELWVVSDGRIAPFHGTFHDYK 708
Query: 1870 AHLKS 1884
L+S
Sbjct: 709 KLLQS 713
>sptr|Q8H0V6|Q8H0V6 ABC transporter protein, putative.
Length = 715
Score = 425 bits (1093), Expect = e-117
Identities = 232/545 (42%), Positives = 334/545 (61%), Gaps = 22/545 (4%)
Frame = +1
Query: 316 RDIHIESLSLTFHGHDLIVDSDLEXXXXXXXXXXXXXXCGKSTLLTAIGCRELP-IPEHM 492
RDIH+++ +++ G DLIVD + GK+T L + + IP +
Sbjct: 173 RDIHMDNFNVSVGGRDLIVDGSITLSFGRHYGLVGRNGTGKTTFLRYMAMHAIEGIPTNC 232
Query: 493 DIYHLSHEIEASDMSALQAIVSCDEERVKL-EKEAEILAAQ------------------- 612
I H+ E+ +ALQ +++ D ER KL E+E +ILA Q
Sbjct: 233 QILHVEQEVVGDKTTALQCVLNTDIERTKLLEEEIQILAKQRETEEPTAKDGMPTKDTVE 292
Query: 613 DDGGGEALERVYERLDAMDAGTAEKRAAEILFGLGFDKQMQAKKTKDFSGGWRMRIALAR 792
D + LE +Y+RLDA+DA TAE RAA IL GL F +MQ K T FSGGWRMRIALAR
Sbjct: 293 GDLMSQRLEEIYKRLDAIDAYTAEARAASILAGLSFTPEMQLKATNTFSGGWRMRIALAR 352
Query: 793 ALFMNPTILLLDEPTNHLDLEACVWLEETLKKFDRILVVISHSQDFLNGVCTNIIHMQNK 972
ALF+ P +LLLDEPTNHLDL A +WLE L K+ + +V+SH+++FLN V T+IIH+QN+
Sbjct: 353 ALFIEPDLLLLDEPTNHLDLHAVLWLETYLTKWPKTFIVVSHAREFLNTVVTDIIHLQNQ 412
Query: 973 KLKLYTGNYDQYVQTRSDLEENQMKQYKWEQEQIASMKEYIARFGHGSAKLARQAQSKEK 1152
KL Y GNYD + +TR + +NQ K ++ + + M+ +I +F + +AK A QS+ K
Sbjct: 413 KLSTYKGNYDIFERTREEQVKNQQKAFESSERSRSHMQAFIDKFRY-NAKRASLVQSRIK 471
Query: 1153 TLAKMERGGLTEKVVRDRVLVFRF-TDVGKLPPPVLQFVEVKFGYTPDNLIYKSLDFGVD 1329
L ++ ++V+ D F F T K PP++ F + FGY L++++L+FG+D
Sbjct: 472 ALDRLAH---VDQVINDPDYKFEFPTPDDKPGPPIISFSDASFGYPGGPLLFRNLNFGID 528
Query: 1330 LDSRIALVGPNGAGKSTLLKLMTGDLAPLDGMVRRHNHLRIAQYHQHLADKLDLDMPALA 1509
LDSRIA+VGPNG GKST+LKL++GDL P G V R +R+A + QH D LDL L
Sbjct: 529 LDSRIAMVGPNGIGKSTILKLISGDLQPSSGTVFRSAKVRVAVFSQHHVDGLDLSSNPLL 588
Query: 1510 YMMKEYPGTEEEKMRAAVGRFGLSGKAQVMPMRNLSDGQRSRVIFAWLAYRQPQLLLLDE 1689
YMM+ YPG E+K+R+ +G G++G + PM LS GQ+SRV FA + +++P LLLLDE
Sbjct: 589 YMMRCYPGVPEQKLRSHLGSLGVTGNLALQPMYTLSGGQKSRVAFAKITFKKPHLLLLDE 648
Query: 1690 PTNHLDIETIDSLAEALNEWDGGLVLVSHDFRLINQVAQEIWVCENQAVTRWEGDIMDFK 1869
P+NHLD++ +++L + L + GG+ +VSHD LI+ E+WV + + + G D+K
Sbjct: 649 PSNHLDLDAVEALIQGLVLFQGGICMVSHDEHLISGSVDELWVVSDGRIAPFHGTFHDYK 708
Query: 1870 AHLKS 1884
L+S
Sbjct: 709 KLLQS 713
>sptr|Q7ZX02|Q7ZX02 Hypothetical protein (Fragment).
Length = 714
Score = 417 bits (1071), Expect = e-115
Identities = 223/532 (41%), Positives = 332/532 (62%), Gaps = 7/532 (1%)
Frame = +1
Query: 313 SRDIHIESLSLTFHGHDLIVDSDLEXXXXXXXXXXXXXXCGKSTLLTAIGCRELPIPEHM 492
S DI IE+ ++F L+ ++L GK+TLL + R L +P H+
Sbjct: 180 SYDIRIENFDVSFGERVLLTGAELHLASGRRYGLVGRNGLGKTTLLKMLAGRSLRVPSHI 239
Query: 493 DIYHLSHEIEASDMSALQAIVSCDEERVKL---EKE--AEILAAQDDGGGEA-LERVYER 654
I H+ E+ D ALQ+++ CD R L EKE A+I A + DG + L +Y +
Sbjct: 240 SILHVEQEVAGDDTPALQSVLECDTLRESLLQEEKELNAKISAGRGDGSESSRLSEIYSK 299
Query: 655 LDAMDAGTAEKRAAEILFGLGFDKQMQAKKTKDFSGGWRMRIALARALFMNPTILLLDEP 834
L+ ++A A RA+ IL GLGF MQ + TK+FSGGWRMR+ALARALF P +LLLDEP
Sbjct: 300 LEEIEADKAPARASVILAGLGFKHTMQQQLTKEFSGGWRMRLALARALFARPDLLLLDEP 359
Query: 835 TNHLDLEACVWLEETLKKFDRILVVISHSQDFLNGVCTNIIHMQNKKLKLYTGNYDQYVQ 1014
TN LD+ A +WLE L+ + ++V+SH ++FLN V T+I+H+ +++L+ Y GN++ +++
Sbjct: 360 TNMLDVRAILWLESYLQTWPSTILVVSHDRNFLNAVATDIVHLHSQRLEAYRGNFESFLK 419
Query: 1015 TRSDLEENQMKQYKWEQEQIASMKEYIARFGHGSAKLARQAQSKEKTLAKMERGGLTEKV 1194
T+ + +NQ ++Y+ + + ++ +I RF + +A A Q QSK K L K+ EK
Sbjct: 420 TKEERLKNQQREYEAQHQYREHIQVFIDRFRY-NANRASQVQSKLKLLEKLPELKPMEK- 477
Query: 1195 VRDRVLVFRFTD-VGKLPPPVLQFVEVKFGYTPDNLIYKSLDFGVDLDSRIALVGPNGAG 1371
D ++ RF D K PP+LQ E+ F Y+ D I+K L DL+SRI +VG NGAG
Sbjct: 478 --DTEVILRFPDGFEKFSPPILQLDEIDFWYSSDQPIFKKLSVSADLESRICVVGENGAG 535
Query: 1372 KSTLLKLMTGDLAPLDGMVRRHNHLRIAQYHQHLADKLDLDMPALAYMMKEYPGTEEEKM 1551
KST+LKL+ G+L+ + G+ H +L+I + QH D+LDL++ A+ + K +PG EE+
Sbjct: 536 KSTMLKLLMGELSAVQGIRNAHRNLKIGYFSQHHIDQLDLNISAVELLAKRFPGKPEEEY 595
Query: 1552 RAAVGRFGLSGKAQVMPMRNLSDGQRSRVIFAWLAYRQPQLLLLDEPTNHLDIETIDSLA 1731
R +G +G+SG+ V P+ +LS GQ+SRV FA + P +LDEPTNHLD+ETI++L
Sbjct: 596 RHQLGSYGISGELAVRPVASLSGGQKSRVAFAQMTMPCPNFYILDEPTNHLDMETIEALG 655
Query: 1732 EALNEWDGGLVLVSHDFRLINQVAQEIWVCENQAVTRWEGDIMDFKAHLKSK 1887
ALN++ GG++LVSHD R I V QE+WVCEN VTR EG +++ L+ +
Sbjct: 656 RALNKFKGGVILVSHDERFIRLVCQELWVCENGGVTRIEGGFDEYRNILQEQ 707
>sptr|Q9M1H3|Q9M1H3 ABC transporter-like protein (At3g54540).
Length = 723
Score = 415 bits (1067), Expect = e-114
Identities = 232/561 (41%), Positives = 343/561 (61%), Gaps = 39/561 (6%)
Frame = +1
Query: 316 RDIHIESLSLTFHGHDLIVDSDLEXXXXXXXXXXXXXXCGKSTLLTAIGCRELPIPEHMD 495
+DI IES S++ G +L+ ++ + GKSTLL + R++P+P+++D
Sbjct: 161 KDITIESFSVSARGKELLKNASVRISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNID 220
Query: 496 IYHLSHEIEASDMSALQAIVSCDEERVKLEKEAEIL-------------AAQDDGGGEAL 636
+ + E+ + SAL A+VS +EE VKL +EAE L DD GE L
Sbjct: 221 VLLVEQEVVGDEKSALNAVVSANEELVKLREEAEALQKSSSGADGENVDGEDDDDTGEKL 280
Query: 637 ERVYERLDAMDAGTAEKRAAEILFGLGFDKQMQAKKTKDFSGGWRMRIALARALFMNPTI 816
+Y+RL + + AE +A++IL GLGF K MQ + T+ FSGGWRMRI+LARALF+ PT+
Sbjct: 281 AELYDRLQILGSDAAEAQASKILAGLGFTKDMQVRATQSFSGGWRMRISLARALFVQPTL 340
Query: 817 LLLDEPTNHLDLEACVWLEETLKKFDRILVVISHSQDFLNGVCTNIIHMQNKKLKLYTGN 996
LLLDEPTNHLDL A +WLEE L ++ + LVV+SH +DFLN VCT IIH+ ++ L Y GN
Sbjct: 341 LLLDEPTNHLDLRAVLWLEEYLCRWKKTLVVVSHDRDFLNTVCTEIIHLHDQNLHFYRGN 400
Query: 997 YDQY----------VQTRSDLEENQMKQYK-----WEQEQIASMKEYIARFGHGSAKLAR 1131
+D + + + D+ + QMK K +QE++ ++ A AK A
Sbjct: 401 FDGFESGYEQRRKEMNKKFDVYDKQMKAAKRTGNRGQQEKVKDRAKFTA------AKEAS 454
Query: 1132 QAQSKEKTLAKMERGGLTEKVVRDRVLVFRFTDVGKLPPPVLQFVEVKFGY--TPDNLIY 1305
+++SK KT+ + + RD +VF F + +L PP+LQ +EV F Y PD +
Sbjct: 455 KSKSKGKTVDEEGPAPEAPRKWRDYSVVFHFPEPTELTPPLLQLIEVSFSYPNRPDFRL- 513
Query: 1306 KSLDFGVDLDSRIALVGPNGAGKSTLLKLMTGDLAPLDGMVRRHNHLRIAQYHQHLADKL 1485
++D G+D+ +R+A+VGPNGAGKSTLL L+ GDL P +G +RR LRI +Y QH D L
Sbjct: 514 SNVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPTEGEMRRSQKLRIGRYSQHFVDLL 573
Query: 1486 DLDMPALAYMMKEYPGTE----EEKMRAAVGRFGLSGKAQVMPMRNLSDGQRSRVIFAWL 1653
+ + Y+++ +P E +E +RA +G+FGL + P+ LS GQ++RV+F +
Sbjct: 574 TMGETPVQYLLRLHPDQEGFSKQEAVRAKLGKFGLPSHNHLSPIAKLSGGQKARVVFTSI 633
Query: 1654 AYRQPQLLLLDEPTNHLDIETIDSLAEALNEWDGGLVLVSHDFRLINQVAQE-----IWV 1818
+ +P +LLLDEPTNHLD+++ID+LA+AL+E+ GG+VLVSHD RLI++V E IWV
Sbjct: 634 SMSKPHILLLDEPTNHLDMQSIDALADALDEFTGGVVLVSHDSRLISRVCAEEEKSQIWV 693
Query: 1819 CENQAVTRWEGDIMDFKAHLK 1881
E+ V + G ++K L+
Sbjct: 694 VEDGTVNFFPGTFEEYKEDLQ 714
>sptr|Q8LFD8|Q8LFD8 Putative ABC transporter.
Length = 723
Score = 415 bits (1066), Expect = e-114
Identities = 232/561 (41%), Positives = 343/561 (61%), Gaps = 39/561 (6%)
Frame = +1
Query: 316 RDIHIESLSLTFHGHDLIVDSDLEXXXXXXXXXXXXXXCGKSTLLTAIGCRELPIPEHMD 495
+DI IES S++ G +L+ ++ + GKSTLL + R++P+P+++D
Sbjct: 161 KDITIESFSVSARGKELLKNASVRISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNID 220
Query: 496 IYHLSHEIEASDMSALQAIVSCDEERVKLEKEAEIL-------------AAQDDGGGEAL 636
+ + E+ + SAL A+VS +EE VKL +EAE L DD GE L
Sbjct: 221 VLLVEQEVVGDEKSALNAVVSANEELVKLREEAEALQKSSSGADGENVDGEDDDDTGEKL 280
Query: 637 ERVYERLDAMDAGTAEKRAAEILFGLGFDKQMQAKKTKDFSGGWRMRIALARALFMNPTI 816
+Y+RL + + AE +A++IL GLGF K MQ + T+ FSGGWRMRI+LARALF+ PT+
Sbjct: 281 AELYDRLQILGSDAAEAQASKILAGLGFTKDMQVRATQSFSGGWRMRISLARALFVQPTL 340
Query: 817 LLLDEPTNHLDLEACVWLEETLKKFDRILVVISHSQDFLNGVCTNIIHMQNKKLKLYTGN 996
LLLDEPTNHLDL A +WLEE L ++ + LVV+SH +DFLN VCT IIH+ ++ L Y GN
Sbjct: 341 LLLDEPTNHLDLRAVLWLEEYLCRWKKTLVVVSHDRDFLNTVCTEIIHLHDQNLHFYRGN 400
Query: 997 YDQY----------VQTRSDLEENQMKQYK-----WEQEQIASMKEYIARFGHGSAKLAR 1131
+D + + + D+ + QMK K +QE++ ++ A AK A
Sbjct: 401 FDGFESGYEQRRKEMNKKFDVYDKQMKAAKRTGNRGQQEKVKDRAKFTA------AKEAS 454
Query: 1132 QAQSKEKTLAKMERGGLTEKVVRDRVLVFRFTDVGKLPPPVLQFVEVKFGY--TPDNLIY 1305
+++SK KT+ + + RD +VF F + +L PP+LQ +EV F Y PD +
Sbjct: 455 KSKSKGKTVDEEGPAPEAPRKWRDYSVVFHFPEPTELTPPLLQLIEVSFSYPNRPDFRL- 513
Query: 1306 KSLDFGVDLDSRIALVGPNGAGKSTLLKLMTGDLAPLDGMVRRHNHLRIAQYHQHLADKL 1485
++D G+D+ +R+A+VGPNGAGKSTLL L+ GDL P +G +RR LRI +Y QH D L
Sbjct: 514 SNVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPTEGEMRRSQKLRIGRYSQHFVDLL 573
Query: 1486 DLDMPALAYMMKEYPGTE----EEKMRAAVGRFGLSGKAQVMPMRNLSDGQRSRVIFAWL 1653
+ + Y+++ +P E +E +RA +G+FGL + P+ LS GQ++RV+F +
Sbjct: 574 TMGETPVQYLLRLHPDQEGFSKQEAVRAKLGKFGLPSHNHLSPIAKLSRGQKARVVFTSI 633
Query: 1654 AYRQPQLLLLDEPTNHLDIETIDSLAEALNEWDGGLVLVSHDFRLINQVAQE-----IWV 1818
+ +P +LLLDEPTNHLD+++ID+LA+AL+E+ GG+VLVSHD RLI++V E IWV
Sbjct: 634 SMSKPHILLLDEPTNHLDMQSIDALADALDEFTGGVVLVSHDSRLISRVCAEEEKSQIWV 693
Query: 1819 CENQAVTRWEGDIMDFKAHLK 1881
E+ V + G ++K L+
Sbjct: 694 VEDGTVNFFPGTFEEYKEDLQ 714
>sptr|Q9N6L4|Q9N6L4 ABC transporter protein 1.
Length = 724
Score = 412 bits (1059), Expect = e-113
Identities = 216/524 (41%), Positives = 323/524 (61%), Gaps = 2/524 (0%)
Frame = +1
Query: 313 SRDIHIESLSLTFHGHDLIVDSDLEXXXXXXXXXXXXXXCGKSTLLTAIGCRELP-IPEH 489
S DI E++ + L+ D+DL GK+TLL A+ REL +
Sbjct: 200 STDIRCEAIHIQLGKQVLLDDTDLVLLTGHKYGLIGRNGAGKTTLLRALAERELEGVSPF 259
Query: 490 MDIYHLSHEIEASDMSALQAIVSCDEERVKL-EKEAEILAAQDDGGGEALERVYERLDAM 666
M I H+ EI A + L+ +++ D ER +L +E E+L D L VY RLDA+
Sbjct: 260 MQILHVEQEIVAGAETPLEVLLATDVERAQLLHEEQELLKQNDTEANTRLNEVYARLDAI 319
Query: 667 DAGTAEKRAAEILFGLGFDKQMQAKKTKDFSGGWRMRIALARALFMNPTILLLDEPTNHL 846
DA +AE RAA IL GL F + M TK SGGWRMR+ALARALF+ P +LLLDEPTNHL
Sbjct: 320 DAHSAEARAATILHGLSFTQDMMTSPTKQLSGGWRMRVALARALFVEPDVLLLDEPTNHL 379
Query: 847 DLEACVWLEETLKKFDRILVVISHSQDFLNGVCTNIIHMQNKKLKLYTGNYDQYVQTRSD 1026
DL A +WLE+ L+ + + L+V+SHS+ FLN VC+ IIH+ L Y GNYDQ+ TR +
Sbjct: 380 DLFAVLWLEQFLRDWQKTLIVVSHSRTFLNNVCSEIIHLIGHHLHYYNGNYDQFEITRVE 439
Query: 1027 LEENQMKQYKWEQEQIASMKEYIARFGHGSAKLARQAQSKEKTLAKMERGGLTEKVVRDR 1206
E Q KQ+ +++Q A ++ +I +F + +A A+ AQS+ K L +ME + VV D
Sbjct: 440 QERQQKKQHAAQEKQRAHIQAFIDKFRY-NANRAKMAQSRIKALERME---MVADVVTDP 495
Query: 1207 VLVFRFTDVGKLPPPVLQFVEVKFGYTPDNLIYKSLDFGVDLDSRIALVGPNGAGKSTLL 1386
F F D + ++ V+ +FGY P ++K ++ G+D SRI LVG NG GKST +
Sbjct: 496 QFAFTFPDAEPVSGSFIEMVDCEFGYKPGVSLFKDVNMGIDESSRIVLVGANGVGKSTFM 555
Query: 1387 KLMTGDLAPLDGMVRRHNHLRIAQYHQHLADKLDLDMPALAYMMKEYPGTEEEKMRAAVG 1566
+ TG L P G V R+ +R+A + QH + L + +L ++ ++P E++++RA +G
Sbjct: 556 NVCTGALEPRSGTVVRNKKIRVAHFAQHNMESLTPQLSSLEFLRSKFPHMEDQQLRAHLG 615
Query: 1567 RFGLSGKAQVMPMRNLSDGQRSRVIFAWLAYRQPQLLLLDEPTNHLDIETIDSLAEALNE 1746
GLSG+ + P+ LS GQ+SR++ AW+ +++P LLLLDEPTNHLDI+T+++L EAL
Sbjct: 616 SMGLSGERALQPIYTLSGGQKSRLVLAWITFQKPHLLLLDEPTNHLDIDTVNALIEALLA 675
Query: 1747 WDGGLVLVSHDFRLINQVAQEIWVCENQAVTRWEGDIMDFKAHL 1878
++GGL+++SHD I + +I+VCE+ V +++GD +++ H+
Sbjct: 676 YNGGLLVISHDEYFITSLCDDIFVCEDNTVKKFDGDFAEYRKHV 719
>sptr|O59672|O59672 Putative amino acid starvation response, yeast gcn
protein kinase activator homolog, non-transporter (ABC)
superfamily.
Length = 736
Score = 410 bits (1054), Expect = e-113
Identities = 222/548 (40%), Positives = 330/548 (60%), Gaps = 29/548 (5%)
Frame = +1
Query: 313 SRDIHIESLSLTFHGHDLIVDSDLEXXXXXXXXXXXXXXCGKSTLLTAIGCRELPIPEHM 492
S+DI I+ + L F GH ++ + L GKSTLL A+ RE+ IP H+
Sbjct: 180 SKDIKIDGIDLAFAGHRILTGASLTLAQGRRYGLTGRNGIGKSTLLRALSRREIAIPTHI 239
Query: 493 DIYHLSHEIEASDMSALQAIVSCD---------EERVK-----LEKEAEILAAQDDGGGE 630
I H+ E+ D ALQ+++ D +E++ +EKE E L+ +D +
Sbjct: 240 TILHVEQEMTGDDTPALQSVLDADVWRKYLIQDQEKITNRLSTIEKELEELS-KDQTADQ 298
Query: 631 ALERVYER---------------LDAMDAGTAEKRAAEILFGLGFDKQMQAKKTKDFSGG 765
A+ R ER L MD+ AE RAA IL GLGF ++MQ+ TK FSGG
Sbjct: 299 AISRRLERERDELDLRLLDIQNKLSEMDSDRAESRAATILAGLGFTQEMQSHATKTFSGG 358
Query: 766 WRMRIALARALFMNPTILLLDEPTNHLDLEACVWLEETLKKFDRILVVISHSQDFLNGVC 945
WRMR++LARALF P +LLLDEP+N LD+ + +L E L+ + I++V+SH + FLN V
Sbjct: 359 WRMRLSLARALFCQPDLLLLDEPSNMLDVPSIAFLSEYLQTYKNIVLVVSHDRSFLNEVA 418
Query: 946 TNIIHMQNKKLKLYTGNYDQYVQTRSDLEENQMKQYKWEQEQIASMKEYIARFGHGSAKL 1125
T+IIH +++L Y GN+ Q+ TR + +NQ+++Y+ + E ++ +I +F + +AK
Sbjct: 419 TDIIHQHSERLDYYKGNFSQFYATREERCKNQLREYEKQMEYRKHLQSFIDKFRYNAAK- 477
Query: 1126 ARQAQSKEKTLAKMERGGLTEKVVRDRVLVFRFTDVGKLPPPVLQFVEVKFGYTPDNLIY 1305
+ +AQS+ K L K+ + EK + + F F V K+ PP+LQ +V F Y P + I
Sbjct: 478 SSEAQSRIKKLEKLP---ILEKPQTEEEVEFEFPPVEKISPPILQMSDVNFEYVPGHPIL 534
Query: 1306 KSLDFGVDLDSRIALVGPNGAGKSTLLKLMTGDLAPLDGMVRRHNHLRIAQYHQHLADKL 1485
K +D V +DSRI +VGPNGAGKST+LKL+ L P G+V RH LRIA + QH D L
Sbjct: 535 KHVDIDVQMDSRIGVVGPNGAGKSTMLKLLIEQLHPTSGIVSRHPRLRIAYFAQHHVDTL 594
Query: 1486 DLDMPALAYMMKEYPGTEEEKMRAAVGRFGLSGKAQVMPMRNLSDGQRSRVIFAWLAYRQ 1665
DL++ AL+++ K +PG EE+ R +G FG+SG + M LS GQ+SRV FA L +
Sbjct: 595 DLNLNALSFLAKTFPGKGEEEYRRHLGAFGVSGPLALQKMITLSGGQKSRVAFACLGLQN 654
Query: 1666 PQLLLLDEPTNHLDIETIDSLAEALNEWDGGLVLVSHDFRLINQVAQEIWVCENQAVTRW 1845
P +L+LDEPTNHLD+E++D+L A+ + GG++LVSHD +++ IW C++ V+++
Sbjct: 655 PHILILDEPTNHLDMESMDALTRAVKRFQGGVILVSHDVDFLDKTCTSIWQCDHNVVSKF 714
Query: 1846 EGDIMDFK 1869
+G I +K
Sbjct: 715 DGTISQYK 722
Score = 65.1 bits (157), Expect = 4e-09
Identities = 41/133 (30%), Positives = 68/133 (51%), Gaps = 1/133 (0%)
Frame = +1
Query: 1477 DKLDLDMPALAYMMKEYPGTEEEKMRAAV-GRFGLSGKAQVMPMRNLSDGQRSRVIFAWL 1653
D+LDL + + + E E A + G + + Q + S G R R+ A
Sbjct: 309 DELDLRLLDIQNKLSEMDSDRAESRAATILAGLGFTQEMQSHATKTFSGGWRMRLSLARA 368
Query: 1654 AYRQPQLLLLDEPTNHLDIETIDSLAEALNEWDGGLVLVSHDFRLINQVAQEIWVCENQA 1833
+ QP LLLLDEP+N LD+ +I L+E L + +++VSHD +N+VA +I ++
Sbjct: 369 LFCQPDLLLLDEPSNMLDVPSIAFLSEYLQTYKNIVLVVSHDRSFLNEVATDIIHQHSER 428
Query: 1834 VTRWEGDIMDFKA 1872
+ ++G+ F A
Sbjct: 429 LDYYKGNFSQFYA 441
>sptrnew|CAE85618|CAE85618 Probable positive effector protein GCN20.
Length = 749
Score = 408 bits (1049), Expect = e-112
Identities = 222/551 (40%), Positives = 337/551 (61%), Gaps = 25/551 (4%)
Frame = +1
Query: 313 SRDIHIESLSLTFHGHDLIVDSDLEXXXXXXXXXXXXXXCGKSTLLTAIGCRELPIPEHM 492
++DI I+++ +T G ++ D+ L GKSTLL A+ RE+PIP H+
Sbjct: 194 TKDIKIDNIDVTIGGLRILTDTTLTLAYGHRYGLVGHNGVGKSTLLRALSRREVPIPTHI 253
Query: 493 DIYHLSHEIEASDMSALQAIVSCDEER-VKLEKEAEIL-------------------AAQ 612
I H+ E+ D A+QA++ D R V L+++AEI+ AA+
Sbjct: 254 SILHVEQELTGDDTPAIQAVLDADVWRKVLLKEQAEIIKKLADIEAQRVGLADTSADAAK 313
Query: 613 DDGGGEA----LERVYERLDAMDAGTAEKRAAEILFGLGFDKQMQAKKTKDFSGGWRMRI 780
D E L + +L M++ AE RAA IL GLGF + Q TK FSGGWRMR+
Sbjct: 314 LDREREVQDNKLGEIQGKLAEMESDKAESRAASILAGLGFSPERQQFATKTFSGGWRMRL 373
Query: 781 ALARALFMNPTILLLDEPTNHLDLEACVWLEETLKKFDRILVVISHSQDFLNGVCTNIIH 960
ALARALF P +LLLDEP+N LD+ + +L L+ + ++V+SH + FLN V T+IIH
Sbjct: 374 ALARALFCEPDLLLLDEPSNMLDVPSITFLSNYLQTYPSTVLVVSHDRAFLNEVATDIIH 433
Query: 961 MQNKKLKLYTG-NYDQYVQTRSDLEENQMKQYKWEQEQIASMKEYIARFGHGSAKLARQA 1137
+ +L Y G N+D + TR + ++ ++Y+ + Q A ++ +I +F + +AK + +A
Sbjct: 434 QHSMRLDYYRGANFDSFYATREERKKVAKREYENQMAQRAHLQAFIDKFRYNAAK-SSEA 492
Query: 1138 QSKEKTLAKMERGGLTEKVVRDRVLVFRFTDVGKLPPPVLQFVEVKFGYTPDNLIYKSLD 1317
QS+ K L KM + E + + F+F +V KL PP++Q EV FGY+PD ++ +++D
Sbjct: 493 QSRIKKLEKMP---VLEPPETEYSVHFKFPEVEKLSPPIVQMSEVTFGYSPDKILLRNVD 549
Query: 1318 FGVDLDSRIALVGPNGAGKSTLLKLMTGDLAPLDGMVRRHNHLRIAQYHQHLADKLDLDM 1497
V LDSRI +VGPNGAGK+T+LKL+ G L+P G++ H LRI + QH D LDL+
Sbjct: 550 LDVQLDSRIGIVGPNGAGKTTILKLLVGKLSPTSGLITMHPRLRIGFFAQHHVDALDLNA 609
Query: 1498 PALAYMMKEYPGTEEEKMRAAVGRFGLSGKAQVMPMRNLSDGQRSRVIFAWLAYRQPQLL 1677
A+++M K YPG +E+ R +G FG++G + M LS GQ+SRV FA LA QP +L
Sbjct: 610 SAVSFMAKTYPGKTDEEYRRQLGAFGITGTTGLQKMALLSGGQKSRVAFACLALTQPHIL 669
Query: 1678 LLDEPTNHLDIETIDSLAEALNEWDGGLVLVSHDFRLINQVAQEIWVCENQAVTRWEGDI 1857
+LDEP+NHLDIE +D+L+EAL +++GG+++VSHD ++ V + +WVCEN V ++ GD+
Sbjct: 670 VLDEPSNHLDIEAMDALSEALQKFEGGVLMVSHDVTMLQTVCKSLWVCENGTVEKFPGDV 729
Query: 1858 MDFKAHLKSKA 1890
+K + ++A
Sbjct: 730 QAYKKRIAAQA 740
>sptr|Q7RZG5|Q7RZG5 Hypothetical protein.
Length = 749
Score = 408 bits (1049), Expect = e-112
Identities = 222/551 (40%), Positives = 337/551 (61%), Gaps = 25/551 (4%)
Frame = +1
Query: 313 SRDIHIESLSLTFHGHDLIVDSDLEXXXXXXXXXXXXXXCGKSTLLTAIGCRELPIPEHM 492
++DI I+++ +T G ++ D+ L GKSTLL A+ RE+PIP H+
Sbjct: 194 TKDIKIDNIDVTIGGLRILTDTTLTLAYGHRYGLVGHNGVGKSTLLRALSRREVPIPTHI 253
Query: 493 DIYHLSHEIEASDMSALQAIVSCDEER-VKLEKEAEIL-------------------AAQ 612
I H+ E+ D A+QA++ D R V L+++AEI+ AA+
Sbjct: 254 SILHVEQELTGDDTPAIQAVLDADVWRKVLLKEQAEIIKKLADIEAQRVGLADTSADAAK 313
Query: 613 DDGGGEA----LERVYERLDAMDAGTAEKRAAEILFGLGFDKQMQAKKTKDFSGGWRMRI 780
D E L + +L M++ AE RAA IL GLGF + Q TK FSGGWRMR+
Sbjct: 314 LDREREVQDNKLGEIQGKLAEMESDKAESRAASILAGLGFSPERQQFATKTFSGGWRMRL 373
Query: 781 ALARALFMNPTILLLDEPTNHLDLEACVWLEETLKKFDRILVVISHSQDFLNGVCTNIIH 960
ALARALF P +LLLDEP+N LD+ + +L L+ + ++V+SH + FLN V T+IIH
Sbjct: 374 ALARALFCEPDLLLLDEPSNMLDVPSITFLSNYLQTYPSTVLVVSHDRAFLNEVATDIIH 433
Query: 961 MQNKKLKLYTG-NYDQYVQTRSDLEENQMKQYKWEQEQIASMKEYIARFGHGSAKLARQA 1137
+ +L Y G N+D + TR + ++ ++Y+ + Q A ++ +I +F + +AK + +A
Sbjct: 434 QHSMRLDYYRGANFDSFYATREERKKVAKREYENQMAQRAHLQAFIDKFRYNAAK-SSEA 492
Query: 1138 QSKEKTLAKMERGGLTEKVVRDRVLVFRFTDVGKLPPPVLQFVEVKFGYTPDNLIYKSLD 1317
QS+ K L KM + E + + F+F +V KL PP++Q EV FGY+PD ++ +++D
Sbjct: 493 QSRIKKLEKMP---VLEPPETEYSVHFKFPEVEKLSPPIVQMSEVTFGYSPDKILLRNVD 549
Query: 1318 FGVDLDSRIALVGPNGAGKSTLLKLMTGDLAPLDGMVRRHNHLRIAQYHQHLADKLDLDM 1497
V LDSRI +VGPNGAGK+T+LKL+ G L+P G++ H LRI + QH D LDL+
Sbjct: 550 LDVQLDSRIGIVGPNGAGKTTILKLLVGKLSPTSGLITMHPRLRIGFFAQHHVDALDLNA 609
Query: 1498 PALAYMMKEYPGTEEEKMRAAVGRFGLSGKAQVMPMRNLSDGQRSRVIFAWLAYRQPQLL 1677
A+++M K YPG +E+ R +G FG++G + M LS GQ+SRV FA LA QP +L
Sbjct: 610 SAVSFMAKTYPGKTDEEYRRQLGAFGITGTTGLQKMALLSGGQKSRVAFACLALTQPHIL 669
Query: 1678 LLDEPTNHLDIETIDSLAEALNEWDGGLVLVSHDFRLINQVAQEIWVCENQAVTRWEGDI 1857
+LDEP+NHLDIE +D+L+EAL +++GG+++VSHD ++ V + +WVCEN V ++ GD+
Sbjct: 670 VLDEPSNHLDIEAMDALSEALQKFEGGVLMVSHDVTMLQTVCKSLWVCENGTVEKFPGDV 729
Query: 1858 MDFKAHLKSKA 1890
+K + ++A
Sbjct: 730 QAYKKRIAAQA 740
>sptr|Q8S694|Q8S694 Putative ABC transporter.
Length = 710
Score = 408 bits (1048), Expect = e-112
Identities = 235/548 (42%), Positives = 337/548 (61%), Gaps = 27/548 (4%)
Frame = +1
Query: 316 RDIHIESLSLTFHGHDLIVDSDLEXXXXXXXXXXXXXXCGKSTLLTAIGCRELPIPEHMD 495
+DI +E+ S++ G +L+ ++ L GKSTLL + R++P+P +D
Sbjct: 156 KDIVLENFSVSARGKELLKNASLRISHGRRYGLVGPNGMGKSTLLKLLSWRQVPVPRSID 215
Query: 496 IYHLSHEIEASDMSALQAIVSCDEERVKLEKE-AEILAAQDDGGGEALERVYERLDAMDA 672
+ + EI + SAL+A+V+ DEE L E A++ A+ D E L VYE+L+ D+
Sbjct: 216 VLLVEQEIIGDNRSALEAVVAADEELAALRAEQAKLEASNDADDNERLAEVYEKLNLRDS 275
Query: 673 GTAEKRAAEILFGLGFDKQMQAKKTKDFSGGWRMRIALARALFMNPTILLLDEPTNHLDL 852
A RA++IL GLGFD+ MQA+ TK FSGGWRMRI+LARALFM PT+LLLDEPTNHLDL
Sbjct: 276 DAARARASKILAGLGFDQAMQARSTKSFSGGWRMRISLARALFMQPTLLLLDEPTNHLDL 335
Query: 853 EACVWLEETL-KKFDRILVVISHSQDFLNGVCTNIIHMQNKKLKLYTGNYDQ----YVQT 1017
A +WLE+ L ++ + L+V+SH +DFLN VC IIH+ +K L +Y GN+D Y Q
Sbjct: 336 RAVLWLEQYLCSQWKKTLIVVSHDRDFLNTVCNEIIHLHDKNLHVYRGNFDDFESGYEQK 395
Query: 1018 RSDLE------ENQMKQYKWEQEQIASMKEYIARFGHGSAKLARQAQSKEKTLAK----M 1167
R ++ E QMK K + A K + K A +++ K K +A M
Sbjct: 396 RKEMNRKFEVFEKQMKAAKKTGSKAAQDKVKGQALSKAN-KEAAKSKGKGKNVANDDDDM 454
Query: 1168 ERGGLTEKVVRDRVLVFRFTDVGKLPPPVLQFVEVKFGYTPDNLIYK--SLDFGVDLDSR 1341
+ L +K + D + F F + L PP+LQ +EV F Y P+ +K +D G+D+ +R
Sbjct: 455 KPADLPQKWL-DYKVEFHFPEPTLLTPPLLQLIEVGFSY-PNRPDFKLSGVDVGIDMGTR 512
Query: 1342 IALVGPNGAGKSTLLKLMTGDLAPLDGMVRRHNHLRIAQYHQHLADKLDLDMPALAYMMK 1521
+A+VGPNGAGKSTLL L+ GDL P G VRR LRI +Y QH D L ++ A+ Y+++
Sbjct: 513 VAIVGPNGAGKSTLLNLLAGDLTPTKGEVRRSQKLRIGRYSQHFVDLLTMEENAVQYLLR 572
Query: 1522 EYPGTE----EEKMRAAVGRFGLSGKAQVMPMRNLSDGQRSRVIFAWLAYRQPQLLLLDE 1689
+P E E +RA +G+FGL G + P+ LS GQ++RV+F ++ P +LLLDE
Sbjct: 573 LHPDQEGMSKAEAVRAKLGKFGLPGHNHLTPIVKLSGGQKARVVFTSISMSHPHILLLDE 632
Query: 1690 PTNHLDIETIDSLAEALNEWDGGLVLVSHDFRLINQVA-----QEIWVCENQAVTRWEGD 1854
PTNHLD+++ID+LA+AL+E+ GG+VLVSHD RLI++V EIWV E+ V +++G
Sbjct: 633 PTNHLDMQSIDALADALDEFTGGVVLVSHDSRLISRVCDDEQRSEIWVVEDGTVNKFDGT 692
Query: 1855 IMDFKAHL 1878
D+K L
Sbjct: 693 FEDYKDEL 700
Score = 80.9 bits (198), Expect = 7e-14
Identities = 65/225 (28%), Positives = 100/225 (44%), Gaps = 29/225 (12%)
Frame = +1
Query: 1300 IYKSLDFGVDLDSRIALVGPNGAGKSTLLKLMTGDLAP---------------------L 1416
+ K+ + R LVGPNG GKSTLLKL++ P L
Sbjct: 172 LLKNASLRISHGRRYGLVGPNGMGKSTLLKLLSWRQVPVPRSIDVLLVEQEIIGDNRSAL 231
Query: 1417 DGMVRRHNHL---RIAQYHQHLADKLDLDMPALAYMMKEYPGTEEEKMRAAVGRF----G 1575
+ +V L R Q ++ D D LA + ++ + + RA + G
Sbjct: 232 EAVVAADEELAALRAEQAKLEASNDAD-DNERLAEVYEKLNLRDSDAARARASKILAGLG 290
Query: 1576 LSGKAQVMPMRNLSDGQRSRVIFAWLAYRQPQLLLLDEPTNHLDIETIDSLAEAL-NEWD 1752
Q ++ S G R R+ A + QP LLLLDEPTNHLD+ + L + L ++W
Sbjct: 291 FDQAMQARSTKSFSGGWRMRISLARALFMQPTLLLLDEPTNHLDLRAVLWLEQYLCSQWK 350
Query: 1753 GGLVLVSHDFRLINQVAQEIWVCENQAVTRWEGDIMDFKAHLKSK 1887
L++VSHD +N V EI ++ + + G+ DF++ + K
Sbjct: 351 KTLIVVSHDRDFLNTVCNEIIHLHDKNLHVYRGNFDDFESGYEQK 395
Database: swall
Posted date: Feb 26, 2004 3:00 PM
Number of letters in database: 439,479,560
Number of sequences in database: 1,381,838
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,596,504,737
Number of Sequences: 1381838
Number of extensions: 34885510
Number of successful extensions: 141658
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 106315
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 134774
length of database: 439,479,560
effective HSP length: 130
effective length of database: 259,840,620
effective search space used: 146290269060
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

