BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= QAD12g05.sg.3.2
(900 letters)
Database: swall
1,381,838 sequences; 439,479,560 total letters
Searching..................................................done
Score E
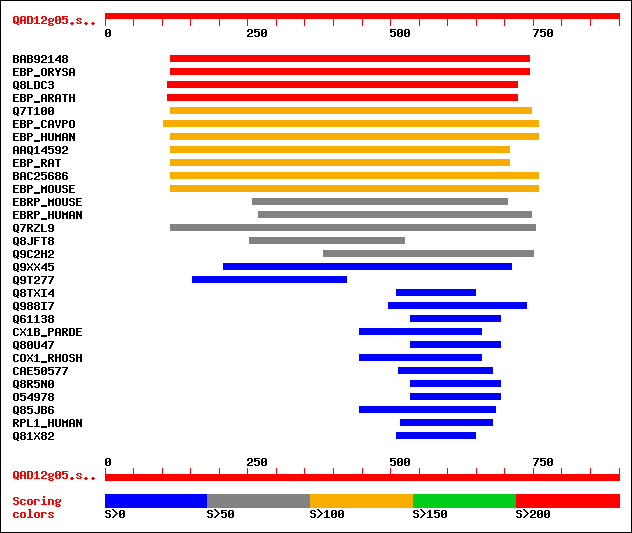
Sequences producing significant alignments: (bits) Value
sptrnew|BAB92148|BAB92148 Putative C-8,7 sterol isomerase. 367 e-100
sw|Q9FTZ2|EBP_ORYSA Probable 3-beta-hydroxysteroid-delta(8),delt... 367 e-100
sptr|Q8LDC3|Q8LDC3 C-8,7 sterol isomerase. 266 2e-70
sw|O48962|EBP_ARATH Probable 3-beta-hydroxysteroid-delta(8),delt... 266 2e-70
sptr|Q7T100|Q7T100 Hypothetical protein. 148 9e-35
sw|Q60490|EBP_CAVPO 3-beta-hydroxysteroid-delta(8),delta(7)-isom... 146 4e-34
sw|Q15125|EBP_HUMAN 3-beta-hydroxysteroid-delta(8),delta(7)-isom... 144 1e-33
sptrnew|AAQ14592|AAQ14592 Sterol 8-isomerase. 144 2e-33
sw|Q9JJ46|EBP_RAT 3-beta-hydroxysteroid-delta(8),delta(7)-isomer... 144 2e-33
sptrnew|BAC25686|BAC25686 18-day embryo whole body cDNA, RIKEN f... 142 9e-33
sw|P70245|EBP_MOUSE 3-beta-hydroxysteroid-delta(8),delta(7)-isom... 142 9e-33
sw|Q9D0P0|EBRP_MOUSE Emopamil binding related protein. 100 3e-20
sw|Q9BY08|EBRP_HUMAN Emopamil binding related protein. 100 5e-20
sptr|Q7RZL9|Q7RZL9 Hypothetical protein. 93 5e-18
sptr|Q8JFT8|Q8JFT8 SI:dZ79F24.4 (Novel protein similar to human ... 66 6e-10
sptr|Q9C2H2|Q9C2H2 (AL513443) related to C-8, 7 sterol isomerase... 59 1e-07
sptr|Q9XX45|Q9XX45 Y38H6C.16 protein. 39 0.080
sptr|Q9T277|Q9T277 NADH dehydrogenase subunit 5 (Fragment). 36 0.68
sptr|Q8TXI4|Q8TXI4 SMC1-family ATPase involved in DNA repair. 35 2.0
sptr|Q988I7|Q988I7 Hypothetical protein mlr6726. 35 2.0
sptr|Q61138|Q61138 Zn-finger protein Pw1. 34 2.6
sw|P98002|CX1B_PARDE Cytochrome c oxidase polypeptide I-beta (EC... 34 2.6
sptr|Q80U47|Q80U47 MKIAA0287 protein (Fragment). 34 2.6
sw|P33517|COX1_RHOSH Cytochrome c oxidase polypeptide I (EC 1.9.... 34 2.6
sptrnew|CAE50577|CAE50577 Putative membrane protein. 34 2.6
sptr|Q8R5N0|Q8R5N0 Zinc finger protein. 33 5.7
sptr|O54978|O54978 Zinc finger protein. 33 5.7
sptr|Q85JB6|Q85JB6 Cytochrome c oxidase subunit 1 (EC 1.9.3.1). 33 5.7
sw|Q8IWN7|RPL1_HUMAN Retinitis pigmentosa 1-like 1 protein. 33 5.7
sptr|Q81X82|Q81X82 Phage major capsid protein, HK97 family. 33 7.5
>sptrnew|BAB92148|BAB92148 Putative C-8,7 sterol isomerase.
Length = 219
Score = 367 bits (942), Expect = e-100
Identities = 174/211 (82%), Positives = 187/211 (88%), Gaps = 1/211 (0%)
Frame = +1
Query: 115 HPYAPAELDLPGFVPLKLSQVEILVSYLGASVFVFLAVWLVSGRCVR-LSKTDRLLMCWW 291
HPYAPAEL LPGFVPL+LSQ +ILV YL S+F+ LAVWL+SGRC R LS TDR LMCWW
Sbjct: 7 HPYAPAELHLPGFVPLQLSQAQILVPYLATSLFLLLAVWLISGRCSRRLSDTDRWLMCWW 66
Query: 292 AFTGLTHIMIEGPFVFTPDFFKKENPNFFDEVWKEYSKGDSRYVARDTATVTVEGITAVL 471
AFTGLTHI+IEG FVF P+FF +NP++FDEVWKEYSKGDSRYVARD ATVTVEGITAVL
Sbjct: 67 AFTGLTHIIIEGTFVFAPNFFSNQNPSYFDEVWKEYSKGDSRYVARDPATVTVEGITAVL 126
Query: 472 EGPASLLAVYAIASRKSFSHILQFAVCLGQLYGCLVYFITAYLDGFNFWVGPFYFWAYFI 651
EGPASLLAVYAIAS KS+SHILQF VCLGQLYGCLVYFITAYLDGFNFW PFYFWAYFI
Sbjct: 127 EGPASLLAVYAIASGKSYSHILQFTVCLGQLYGCLVYFITAYLDGFNFWTSPFYFWAYFI 186
Query: 652 GANSFWIWIPMLIAIRSWKKTCAAFQAEKVK 744
GANS W+ IP +IAIRSWKK CAAFQ EKVK
Sbjct: 187 GANSSWVVIPTMIAIRSWKKICAAFQGEKVK 217
>sw|Q9FTZ2|EBP_ORYSA Probable
3-beta-hydroxysteroid-delta(8),delta(7)-isomerase (EC
5.3.3.5) (Cholestenol delta-isomerase) (Delta8-delta7
sterol isomerase) (D8-D7 sterol isomerase).
Length = 219
Score = 367 bits (942), Expect = e-100
Identities = 174/211 (82%), Positives = 187/211 (88%), Gaps = 1/211 (0%)
Frame = +1
Query: 115 HPYAPAELDLPGFVPLKLSQVEILVSYLGASVFVFLAVWLVSGRCVR-LSKTDRLLMCWW 291
HPYAPAEL LPGFVPL+LSQ +ILV YL S+F+ LAVWL+SGRC R LS TDR LMCWW
Sbjct: 7 HPYAPAELHLPGFVPLQLSQAQILVPYLATSLFLLLAVWLISGRCSRRLSDTDRWLMCWW 66
Query: 292 AFTGLTHIMIEGPFVFTPDFFKKENPNFFDEVWKEYSKGDSRYVARDTATVTVEGITAVL 471
AFTGLTHI+IEG FVF P+FF +NP++FDEVWKEYSKGDSRYVARD ATVTVEGITAVL
Sbjct: 67 AFTGLTHIIIEGTFVFAPNFFSNQNPSYFDEVWKEYSKGDSRYVARDPATVTVEGITAVL 126
Query: 472 EGPASLLAVYAIASRKSFSHILQFAVCLGQLYGCLVYFITAYLDGFNFWVGPFYFWAYFI 651
EGPASLLAVYAIAS KS+SHILQF VCLGQLYGCLVYFITAYLDGFNFW PFYFWAYFI
Sbjct: 127 EGPASLLAVYAIASGKSYSHILQFTVCLGQLYGCLVYFITAYLDGFNFWTSPFYFWAYFI 186
Query: 652 GANSFWIWIPMLIAIRSWKKTCAAFQAEKVK 744
GANS W+ IP +IAIRSWKK CAAFQ EKVK
Sbjct: 187 GANSSWVVIPTMIAIRSWKKICAAFQGEKVK 217
>sptr|Q8LDC3|Q8LDC3 C-8,7 sterol isomerase.
Length = 223
Score = 266 bits (681), Expect = 2e-70
Identities = 122/205 (59%), Positives = 158/205 (77%)
Frame = +1
Query: 109 MGHPYAPAELDLPGFVPLKLSQVEILVSYLGASVFVFLAVWLVSGRCVRLSKTDRLLMCW 288
+ HPY P +L+LPG+VP+ +S I+ YLG+S+ V VWL+ GR + +K D+LLMCW
Sbjct: 4 LAHPYVPRDLNLPGYVPISMSMSSIVSIYLGSSLLVVSLVWLLFGR--KKAKLDKLLMCW 61
Query: 289 WAFTGLTHIMIEGPFVFTPDFFKKENPNFFDEVWKEYSKGDSRYVARDTATVTVEGITAV 468
W FTGLTH+++EG FVF+P+FFK + EVWKEYSKGDSRYV RD+A V+VEGITAV
Sbjct: 62 WTFTGLTHVILEGYFVFSPEFFKDNTSAYLAEVWKEYSKGDSRYVGRDSAVVSVEGITAV 121
Query: 469 LEGPASLLAVYAIASRKSFSHILQFAVCLGQLYGCLVYFITAYLDGFNFWVGPFYFWAYF 648
+ GPASLLA+YAIA KS+S++LQ A+ + QLYGCLVYFITA L+G NF FY+++Y+
Sbjct: 122 IVGPASLLAIYAIAKEKSYSYVLQLAISVCQLYGCLVYFITAILEGDNFATNSFYYYSYY 181
Query: 649 IGANSFWIWIPMLIAIRSWKKTCAA 723
IGAN +W+ IP LI+ R WKK CAA
Sbjct: 182 IGANCWWVLIPSLISFRCWKKICAA 206
>sw|O48962|EBP_ARATH Probable
3-beta-hydroxysteroid-delta(8),delta(7)-isomerase (EC
5.3.3.5) (Cholestenol delta-isomerase) (Delta8-delta7
sterol isomerase) (D8-D7 sterol isomerase).
Length = 223
Score = 266 bits (681), Expect = 2e-70
Identities = 122/205 (59%), Positives = 158/205 (77%)
Frame = +1
Query: 109 MGHPYAPAELDLPGFVPLKLSQVEILVSYLGASVFVFLAVWLVSGRCVRLSKTDRLLMCW 288
+ HPY P +L+LPG+VP+ +S I+ YLG+S+ V VWL+ GR + +K D+LLMCW
Sbjct: 4 LAHPYVPRDLNLPGYVPISMSMSSIVSIYLGSSLLVVSLVWLLFGR--KKAKLDKLLMCW 61
Query: 289 WAFTGLTHIMIEGPFVFTPDFFKKENPNFFDEVWKEYSKGDSRYVARDTATVTVEGITAV 468
W FTGLTH+++EG FVF+P+FFK + EVWKEYSKGDSRYV RD+A V+VEGITAV
Sbjct: 62 WTFTGLTHVILEGYFVFSPEFFKDNTSAYLAEVWKEYSKGDSRYVGRDSAVVSVEGITAV 121
Query: 469 LEGPASLLAVYAIASRKSFSHILQFAVCLGQLYGCLVYFITAYLDGFNFWVGPFYFWAYF 648
+ GPASLLA+YAIA KS+S++LQ A+ + QLYGCLVYFITA L+G NF FY+++Y+
Sbjct: 122 IVGPASLLAIYAIAKEKSYSYVLQLAISVCQLYGCLVYFITAILEGDNFATNSFYYYSYY 181
Query: 649 IGANSFWIWIPMLIAIRSWKKTCAA 723
IGAN +W+ IP LI+ R WKK CAA
Sbjct: 182 IGANCWWVLIPSLISFRCWKKICAA 206
>sptr|Q7T100|Q7T100 Hypothetical protein.
Length = 229
Score = 148 bits (374), Expect = 9e-35
Identities = 84/220 (38%), Positives = 116/220 (52%), Gaps = 9/220 (4%)
Frame = +1
Query: 115 HPYAPAELDLPGFVPLKLSQVEILVSYLGASVFVFLAVWLVSGRCVRLSKTDRLLMCWWA 294
HPY P EL L G+ P EIL S + A+WL++GR + RL++CW+
Sbjct: 11 HPYWPRELRLEGYRPNDRYVGEILAFLFSVSGVLLAAMWLLTGRARGMGTGRRLVICWFT 70
Query: 295 FTGLTHIMIEGPFVFTPDFFKKENPNFFDEVWKEYSKGDSRYVARDTATVTVEGITAVLE 474
H +IEG F + + F ++WKEYSKGDSRYV+ D TV +E ITA
Sbjct: 71 VCAFIHCVIEGWFSLYHEDLPWDQA-FLSQLWKEYSKGDSRYVSADNFTVCMETITAWAW 129
Query: 475 GPASLLAVYAIASRKSFSHILQFAVCLGQLYGCLVYFITAYLDGF---NFWVGPFYFWAY 645
GP S V + + ++LQ V LGQLYG ++YF T Y +GF W P YFW Y
Sbjct: 130 GPLSAWTVISFLQQNPQRYVLQLIVSLGQLYGDVLYFYTEYREGFRHSEMW-HPLYFWFY 188
Query: 646 FIGANSFWIWIPMLIAIRSW------KKTCAAFQAEKVKK 747
F+ N+ WI IP ++ + SW ++ C A + K K+
Sbjct: 189 FVFMNALWIIIPSILIVDSWLQLSQAQRLCDAAPSSKAKR 228
>sw|Q60490|EBP_CAVPO
3-beta-hydroxysteroid-delta(8),delta(7)-isomerase (EC
5.3.3.5) (Cholestenol delta-isomerase) (Delta8-delta7
sterol isomerase) (D8-D7 sterol isomerase) (Emopamil
binding protein).
Length = 228
Score = 146 bits (369), Expect = 4e-34
Identities = 95/230 (41%), Positives = 123/230 (53%), Gaps = 11/230 (4%)
Frame = +1
Query: 103 TSMG--HPYAPAELDLPGFVPLKLSQ---VEILVSYLGASVFVFLAVWLVSGRC--VRLS 261
TS G HPY P L L FVP LS V +L + GA V + +WL+S R V L
Sbjct: 3 TSTGPLHPYWPRHLRLDHFVPNDLSAWYIVTVLFTVFGALV---VTMWLLSSRASVVPLG 59
Query: 262 KTDRLLMCWWAFTGLTHIMIEGPFVFTPDFFKKENPNFFDEVWKEYSKGDSRYVARDTAT 441
RL +CW+A H++IEG FV + F ++WKEY+KGDSRY+ D
Sbjct: 60 TWRRLSVCWFAVCAFVHLVIEGWFVLYQKAILGDQA-FLSQLWKEYAKGDSRYIIEDNFI 118
Query: 442 VTVEGITAVLEGPASLLAVYAIASRKSFSHILQFAVCLGQLYGCLVYFITAYLDGFNF-- 615
+ +E IT VL GP SL AV A + ++LQF + LGQ+YG L+YF+T Y DGF
Sbjct: 119 ICMESITVVLWGPLSLWAVIAFLRQHPSRYVLQFVISLGQIYGDLLYFLTEYRDGFQHGE 178
Query: 616 WVGPFYFWAYFIGANSFWIWIPMLIAIRSWKKTCAAFQA--EKVKKTK*K 759
P YFW YF N W+ IP ++ S K+ A A KV K+K K
Sbjct: 179 MGHPIYFWFYFFFMNVLWLVIPGVLFFDSVKQFYGAQNALDTKVMKSKGK 228
>sw|Q15125|EBP_HUMAN
3-beta-hydroxysteroid-delta(8),delta(7)-isomerase (EC
5.3.3.5) (Cholestenol delta-isomerase) (Delta8-delta7
sterol isomerase) (D8-D7 sterol isomerase) (Emopamil
binding protein).
Length = 230
Score = 144 bits (364), Expect = 1e-33
Identities = 84/219 (38%), Positives = 114/219 (52%), Gaps = 4/219 (1%)
Frame = +1
Query: 115 HPYAPAELDLPGFVPLKLSQVEILVSYLGASVFVFLAVWLVSGRC--VRLSKTDRLLMCW 288
HPY P L L FVP IL + + + WL+SGR V L RL +CW
Sbjct: 9 HPYWPQHLRLDNFVPNDRPTWHILAGLFSVTGVLVVTTWLLSGRAAVVPLGTWRRLSLCW 68
Query: 289 WAFTGLTHIMIEGPFVFTPDFFKKENPNFFDEVWKEYSKGDSRYVARDTATVTVEGITAV 468
+A G H++IEG FV + + F ++WKEY+KGDSRY+ D TV +E ITA
Sbjct: 69 FAVCGFIHLVIEGWFVLYYEDLLGDQA-FLSQLWKEYAKGDSRYILGDNFTVCMETITAC 127
Query: 469 LEGPASLLAVYAIASRKSFSHILQFAVCLGQLYGCLVYFITAYLDGFNFWV--GPFYFWA 642
L GP SL V A + ILQ V +GQ+YG ++YF+T + DGF P YFW
Sbjct: 128 LWGPLSLWVVIAFLRQHPLRFILQLVVSVGQIYGDVLYFLTEHRDGFQHGELGHPLYFWF 187
Query: 643 YFIGANSFWIWIPMLIAIRSWKKTCAAFQAEKVKKTK*K 759
YF+ N+ W+ +P ++ + + K A K TK K
Sbjct: 188 YFVFMNALWLVLPGVLVLDAVKHLTHAQSTLDAKATKAK 226
>sptrnew|AAQ14592|AAQ14592 Sterol 8-isomerase.
Length = 230
Score = 144 bits (362), Expect = 2e-33
Identities = 81/202 (40%), Positives = 108/202 (53%), Gaps = 4/202 (1%)
Frame = +1
Query: 115 HPYAPAELDLPGFVPLKLSQVEILVSYLGASVFVFLAVWLVSGRC--VRLSKTDRLLMCW 288
HPY P L L FVP L ILV S + + WL+S R V L RL +CW
Sbjct: 9 HPYWPRHLRLDNFVPNDLPTWHILVGLFSFSGVLIVITWLLSSRVSVVPLGTGRRLALCW 68
Query: 289 WAFTGLTHIMIEGPFVFTPDFFKKENPNFFDEVWKEYSKGDSRYVARDTATVTVEGITAV 468
+A H++IEG F F + E+ F ++WKEYSKGDSRY+ D V +E +TA
Sbjct: 69 FAVCTFIHLVIEGWFSFYHEILL-EDQAFLSQLWKEYSKGDSRYILSDGFIVCMESVTAC 127
Query: 469 LEGPASLLAVYAIASRKSFSHILQFAVCLGQLYGCLVYFITAYLDGFNFWV--GPFYFWA 642
L GP SL V A + F +LQ V +GQ+YG ++YF+T DGF P YFW
Sbjct: 128 LWGPLSLWVVIAFLRHQPFRFVLQLVVSVGQIYGDVLYFLTELRDGFQHGELGHPLYFWF 187
Query: 643 YFIGANSFWIWIPMLIAIRSWK 708
YF+ N+ W+ IP ++ + K
Sbjct: 188 YFVIMNAIWLVIPGILVFDAIK 209
>sw|Q9JJ46|EBP_RAT 3-beta-hydroxysteroid-delta(8),delta(7)-isomerase
(EC 5.3.3.5) (Cholestenol delta-isomerase)
(Delta8-delta7 sterol isomerase) (D8-D7 sterol
isomerase) (Emopamil binding protein).
Length = 230
Score = 144 bits (362), Expect = 2e-33
Identities = 81/202 (40%), Positives = 108/202 (53%), Gaps = 4/202 (1%)
Frame = +1
Query: 115 HPYAPAELDLPGFVPLKLSQVEILVSYLGASVFVFLAVWLVSGRC--VRLSKTDRLLMCW 288
HPY P L L FVP L ILV S + + WL+S R V L RL +CW
Sbjct: 9 HPYWPRHLRLDNFVPNDLPTWHILVGLFSFSGVLIVITWLLSSRVSVVPLGTGRRLALCW 68
Query: 289 WAFTGLTHIMIEGPFVFTPDFFKKENPNFFDEVWKEYSKGDSRYVARDTATVTVEGITAV 468
+A H++IEG F F + E+ F ++WKEYSKGDSRY+ D V +E +TA
Sbjct: 69 FAVCTFIHLVIEGWFSFYHEILL-EDQAFLSQLWKEYSKGDSRYILSDGFIVCMESVTAC 127
Query: 469 LEGPASLLAVYAIASRKSFSHILQFAVCLGQLYGCLVYFITAYLDGFNFWV--GPFYFWA 642
L GP SL V A + F +LQ V +GQ+YG ++YF+T DGF P YFW
Sbjct: 128 LWGPLSLWVVIAFLRHQPFRFVLQLVVSVGQIYGDVLYFLTELRDGFQHGELGHPLYFWF 187
Query: 643 YFIGANSFWIWIPMLIAIRSWK 708
YF+ N+ W+ IP ++ + K
Sbjct: 188 YFVIMNAIWLVIPGILVFDAIK 209
>sptrnew|BAC25686|BAC25686 18-day embryo whole body cDNA, RIKEN
full-length enriched library, clone:1110055D13
product:phenylalkylamine Ca2+ antagonist (emopamil)
binding protein, full insert sequence.
Length = 230
Score = 142 bits (357), Expect = 9e-33
Identities = 85/221 (38%), Positives = 117/221 (52%), Gaps = 6/221 (2%)
Frame = +1
Query: 115 HPYAPAELDLPGFVPLKLSQVEILVSYLGASVFVFLAVWLVSGRC--VRLSKTDRLLMCW 288
HPY P L L FVP L ILV S + + WL+S R V L RL +CW
Sbjct: 9 HPYWPRHLKLDNFVPNDLPTSHILVGLFSISGGLIVITWLLSSRASVVPLGAGRRLALCW 68
Query: 289 WAFTGLTHIMIEGPFVFTPDFFKKENPNFFDEVWKEYSKGDSRYVARDTATVTVEGITAV 468
+A H++IEG F E+ F ++WKEYSKGDSRY+ D+ V +E +TA
Sbjct: 69 FAVCTFIHLVIEGWFSLYNGILL-EDQAFLSQLWKEYSKGDSRYILSDSFVVCMETVTAC 127
Query: 469 LEGPASLLAVYAIASRKSFSHILQFAVCLGQLYGCLVYFITAYLDGFNFW-VG-PFYFWA 642
L GP SL V A ++ F +LQ V +GQ+YG ++YF+T +G +G P YFW
Sbjct: 128 LWGPLSLWVVIAFLRQQPFRFVLQLVVSMGQIYGDVLYFLTELHEGLQHGEIGHPVYFWF 187
Query: 643 YFIGANSFWIWIPMLIAIRSWKKTCAAFQA--EKVKKTK*K 759
YF+ N+ W+ IP ++ + + K +A KV K K K
Sbjct: 188 YFVFLNAVWLVIPSILVLDAIKHLTSAQSVLDSKVMKIKSK 228
>sw|P70245|EBP_MOUSE
3-beta-hydroxysteroid-delta(8),delta(7)-isomerase (EC
5.3.3.5) (Cholestenol delta-isomerase) (Delta8-delta7
sterol isomerase) (D8-D7 sterol isomerase) (Emopamil
binding protein).
Length = 230
Score = 142 bits (357), Expect = 9e-33
Identities = 85/221 (38%), Positives = 117/221 (52%), Gaps = 6/221 (2%)
Frame = +1
Query: 115 HPYAPAELDLPGFVPLKLSQVEILVSYLGASVFVFLAVWLVSGRC--VRLSKTDRLLMCW 288
HPY P L L FVP L ILV S + + WL+S R V L RL +CW
Sbjct: 9 HPYWPRHLKLDNFVPNDLPTSHILVGLFSISGGLIVITWLLSSRASVVPLGAGRRLALCW 68
Query: 289 WAFTGLTHIMIEGPFVFTPDFFKKENPNFFDEVWKEYSKGDSRYVARDTATVTVEGITAV 468
+A H++IEG F E+ F ++WKEYSKGDSRY+ D+ V +E +TA
Sbjct: 69 FAVCTFIHLVIEGWFSLYNGILL-EDQAFLSQLWKEYSKGDSRYILSDSFVVCMETVTAC 127
Query: 469 LEGPASLLAVYAIASRKSFSHILQFAVCLGQLYGCLVYFITAYLDGFNFW-VG-PFYFWA 642
L GP SL V A ++ F +LQ V +GQ+YG ++YF+T +G +G P YFW
Sbjct: 128 LWGPLSLWVVIAFLRQQPFRFVLQLVVSMGQIYGDVLYFLTELHEGLQHGEIGHPVYFWF 187
Query: 643 YFIGANSFWIWIPMLIAIRSWKKTCAAFQA--EKVKKTK*K 759
YF+ N+ W+ IP ++ + + K +A KV K K K
Sbjct: 188 YFVFLNAVWLVIPSILVLDAIKHLTSAQSVLDSKVMKIKSK 228
>sw|Q9D0P0|EBRP_MOUSE Emopamil binding related protein.
Length = 206
Score = 100 bits (249), Expect = 3e-20
Identities = 49/155 (31%), Positives = 82/155 (52%), Gaps = 6/155 (3%)
Frame = +1
Query: 259 SKTDRLLMCWWAFTGLTHIMIEGPFVFTPDFFK-KENPNFFDEVWKEYSKGDSRYVARDT 435
S +R ++ W + L H ++EG FV+ ++ +WKEY K D+R++ D
Sbjct: 37 SAVERWVLAWLCYDSLVHFVLEGAFVYLSIVGNVADSQGLIASLWKEYGKADTRWLYSDP 96
Query: 436 ATVTVEGITAVLEGPASLLAVYAIASRKSFSHILQFAVCLGQLYGCLVYFITAYLDG--- 606
V++E +T VL+G +L+ +YAI K + H +Q +C+ +LYGC + F +L G
Sbjct: 97 TVVSLEILTVVLDGLLALVLIYAIVKEKYYRHFVQIVLCVCELYGCWMTFFPEWLVGSPS 156
Query: 607 --FNFWVGPFYFWAYFIGANSFWIWIPMLIAIRSW 705
+ W+ Y W Y + N W+ IP L+ +SW
Sbjct: 157 LNTSSWL---YLWVYLVFFNGLWVLIPGLLLWQSW 188
>sw|Q9BY08|EBRP_HUMAN Emopamil binding related protein.
Length = 206
Score = 99.8 bits (247), Expect = 5e-20
Identities = 57/168 (33%), Positives = 83/168 (49%), Gaps = 8/168 (4%)
Frame = +1
Query: 268 DRLLMCWWAFTGLTHIMIEGPFVFTPDFFKKENPN-FFDEVWKEYSKGDSRYVARDTATV 444
DR + W + L H +EGPFV+ N + +WKEY K D+R+V D V
Sbjct: 40 DRGALIWLCYDALVHFALEGPFVYLSLVGNVANSDGLIASLWKEYGKADARWVYFDPTIV 99
Query: 445 TVEGITAVLEGPASLLAVYAIASRKSFSHILQFAVCLGQLYGCLVYFITAYLD-----GF 609
+VE +T L+G +L +YAI K + H LQ +C+ +LYGC + F+ +L
Sbjct: 100 SVEILTVALDGSLALFLIYAIVKEKYYRHFLQITLCVCELYGCWMTFLPEWLTRSPNLNT 159
Query: 610 NFWVGPFYFWAYFIGANSFWIWIPMLIAIRSWKKTCAAFQAE--KVKK 747
+ W+ Y W Y N W+ IP L+ +SW + Q E VKK
Sbjct: 160 SNWL---YCWLYLFFFNGVWVLIPGLLLWQSWLELKKMHQKETSSVKK 204
>sptr|Q7RZL9|Q7RZL9 Hypothetical protein.
Length = 345
Score = 93.2 bits (230), Expect = 5e-18
Identities = 62/221 (28%), Positives = 103/221 (46%), Gaps = 8/221 (3%)
Frame = +1
Query: 115 HPYAPAELDLPGFVPLKLSQVEILVSYLGASVFVFLAVWLVSGRCVR-LSKTDRLLMCWW 291
HPY P E+ +P + P + +L ++ GA+ + + + + + L+ D ++ W
Sbjct: 22 HPYYPREVVVPNYAPNTYTLPMVLGAFGGATALLMVGTVAIGKKVNKQLTWKDAGVLAW- 80
Query: 292 AFTGLTHIMIEGPFVFTPDFFKKENPNFFDEVWKEYSKGDSRYVARDTATVTVEGITAVL 471
FT +++ D F ++WKEY+ DSRY+ D + +E +T +
Sbjct: 81 -FTLCYYVLNHATLASKQDIFA--------QLWKEYALSDSRYMTSDPFMLCIETLTVLT 131
Query: 472 EGPASLLAVYA-IASRKSFSHILQFAVCLGQLYGCLVYFITAYLD----GFNF-WVGPFY 633
GP S LAV+A I S HI Q V +G LYG +YF T + G ++ Y
Sbjct: 132 WGPLSFLAVFAIIKGNTSLRHITQTIVSVGHLYGVALYFGTCFFQEKFRGISYSRPETLY 191
Query: 634 FWAYFIGANSFWIWIPMLIAIRSWKKTCAAFQA-EKVKKTK 753
+W Y+ G N+ W+ +P ++ +S K KVK K
Sbjct: 192 YWVYYAGMNAPWVIVPAILLFQSSKTISQGLSVLNKVKSMK 232
>sptr|Q8JFT8|Q8JFT8 SI:dZ79F24.4 (Novel protein similar to human
emopamil binding related protein) (Fragment).
Length = 133
Score = 66.2 bits (160), Expect = 6e-10
Identities = 33/92 (35%), Positives = 50/92 (54%), Gaps = 1/92 (1%)
Frame = +1
Query: 253 RLSKTDRLLMCWWAFTGLTHIMIEGPFVFTPDFFK-KENPNFFDEVWKEYSKGDSRYVAR 429
+ S DR ++ W + + H +EGPFV+ + N E+WKEY K D R++
Sbjct: 42 KCSAVDRWVLVWLFYDAIVHFTLEGPFVYMSLVGNVASSDNLLAELWKEYGKADERWLYS 101
Query: 430 DTATVTVEGITAVLEGPASLLAVYAIASRKSF 525
D V++E +T VL+G +LL VYAI K +
Sbjct: 102 DPTIVSLELLTVVLDGSLALLLVYAIIKDKHY 133
>sptr|Q9C2H2|Q9C2H2 (AL513443) related to C-8, 7 sterol
isomerase/emopamil-binding protein (Hypothetical
protein).
Length = 278
Score = 58.5 bits (140), Expect = 1e-07
Identities = 36/132 (27%), Positives = 62/132 (46%), Gaps = 9/132 (6%)
Frame = +1
Query: 382 EVWKEYSKGDSRYVARDTATVTVEGITAVLEGPASLLAVYAIASRKSFSHILQFAVCLGQ 561
E+W Y+K D R+ D + V++E +T ++ GP + Y IA + S +I+ + +
Sbjct: 149 ELWMVYAKADKRWAGVDLSVVSLELLTVLVVGPLACWVCYDIAKKNSRVNIVMIMIATAE 208
Query: 562 LYGCLVYFITAYLDG-----FNFWVGPFYFWAYFIGANSFWIWIPMLIAIRSWKKTCAAF 726
+YG + F +L G + W+ Y W Y N W+ IP + + + AF
Sbjct: 209 IYGGWMTFCPEWLVGSVNLDTSNWM---YLWLYLAFFNGLWVVIPAYVIYVASGEIMGAF 265
Query: 727 Q----AEKVKKT 750
+ A K KK+
Sbjct: 266 KVRDAAAKAKKS 277
>sptr|Q9XX45|Q9XX45 Y38H6C.16 protein.
Length = 244
Score = 39.3 bits (90), Expect = 0.080
Identities = 41/183 (22%), Positives = 68/183 (37%), Gaps = 15/183 (8%)
Frame = +1
Query: 208 VFVFLAVWLVSGRCVRLSKTDRLLMCWWAFTGLTHIM-----IEGPFVFTPDFFKKENPN 372
+F+FL + ++ R + W +G+ ++ + P+ PD F N
Sbjct: 42 IFLFLTIIKLASSMPRPPVLPPWVATWLLVSGVICLVDVIYTMFRPYTNAPDGFVS-NTL 100
Query: 373 FFDEVWKEYSKGDSRYVARDTATVTVEGITAVLEGPASLLAVYAIASRKSFSHILQF--- 543
F+ WK YS D RY G ++E + LAVY R + +L F
Sbjct: 101 FYG--WKLYSSVDIRYADTKDVVTCSTGRVMLIEITLNFLAVYLALKRSRHALLLAFTTS 158
Query: 544 AVCLGQLYGCLVYFI-------TAYLDGFNFWVGPFYFWAYFIGANSFWIWIPMLIAIRS 702
A + + LV +I + + D + + FW N W+ +P L
Sbjct: 159 AFVFWKTFWYLVMYINPPPGTPSFFTDNYGYLGITLIFWI----PNGVWVVMPFLAMCSL 214
Query: 703 WKK 711
W K
Sbjct: 215 WNK 217
>sptr|Q9T277|Q9T277 NADH dehydrogenase subunit 5 (Fragment).
Length = 355
Score = 36.2 bits (82), Expect = 0.68
Identities = 25/93 (26%), Positives = 43/93 (46%), Gaps = 3/93 (3%)
Frame = +1
Query: 154 VPLKLSQVEILVSYLGASVFVFLAVWLVS--GRCVRLSKTDRLLMCWWAFTGL-THIMIE 324
+P+KL + +LVS++GA + L+ + +S ++ K C W + T M
Sbjct: 231 LPIKLKILALLVSFIGAWIGYELSKFSISWFSNSLKFYKYSYFFGCMWFMPNISTFSMNY 290
Query: 325 GPFVFTPDFFKKENPNFFDEVWKEYSKGDSRYV 423
P V + + +K FD+ W EY G Y+
Sbjct: 291 VPLVMSYNLYKN-----FDQGWNEYFGGQGMYM 318
>sptr|Q8TXI4|Q8TXI4 SMC1-family ATPase involved in DNA repair.
Length = 876
Score = 34.7 bits (78), Expect = 2.0
Identities = 19/49 (38%), Positives = 30/49 (61%), Gaps = 3/49 (6%)
Frame = -1
Query: 648 EIRPEVERADPEVEAVQVRGDEVNQASVELAQTDGE---LENVAEGLPG 511
E++ EV+ +PEVE ++ R +E+ +A E + +GE LEN E L G
Sbjct: 210 ELKREVKELEPEVEELKERLNELREAKREFERLEGELRLLENKIESLKG 258
>sptr|Q988I7|Q988I7 Hypothetical protein mlr6726.
Length = 334
Score = 34.7 bits (78), Expect = 2.0
Identities = 22/87 (25%), Positives = 42/87 (48%), Gaps = 6/87 (6%)
Frame = +1
Query: 496 VYAIASRKSFSHILQFAVCLGQLYGCLVYFITAYLDGFNFWVGPF------YFWAYFIGA 657
++AIA S+ L V L + + ++ +L G + + PF +FW++ G
Sbjct: 238 LFAIAQSGSWQLALIVFVGLNVIQFAIGSYLEPFLTGASLAISPFAVMFAVFFWSFMWGV 297
Query: 658 NSFWIWIPMLIAIRSWKKTCAAFQAEK 738
+I +P+LI + +W CA Q+ +
Sbjct: 298 PGAFIGVPILIVVVTW---CAHEQSTR 321
>sptr|Q61138|Q61138 Zn-finger protein Pw1.
Length = 1378
Score = 34.3 bits (77), Expect = 2.6
Identities = 22/53 (41%), Positives = 26/53 (49%)
Frame = -1
Query: 693 GDEHRYPDPEAVCANEIRPEVERADPEVEAVQVRGDEVNQASVELAQTDGELE 535
G +PE A PEVE A+PEVEA + G E E A+ DGE E
Sbjct: 1178 GSNAEAAEPEVEAAE---PEVEAAEPEVEAAEPNG-EAEGPDGEAAEPDGEAE 1226
>sw|P98002|CX1B_PARDE Cytochrome c oxidase polypeptide I-beta (EC
1.9.3.1) (Cytochrome AA3 subunit 1-beta).
Length = 558
Score = 34.3 bits (77), Expect = 2.6
Identities = 24/76 (31%), Positives = 37/76 (48%), Gaps = 4/76 (5%)
Frame = +1
Query: 445 TVEGITAVLEGPASLLAVYAIASRKSFSHILQFAVCLGQLYGCL--VYFITAYLDGFNF- 615
TV G+T V+ A L VY + +H + + LG ++G VY+ + G +
Sbjct: 384 TVGGVTGVVLSQAPLDRVYH-DTYYVVAHF-HYVMSLGAVFGIFAGVYYWIGKMSGRQYP 441
Query: 616 -WVGPFYFWAYFIGAN 660
W G +FW FIG+N
Sbjct: 442 EWAGQLHFWMMFIGSN 457
>sptr|Q80U47|Q80U47 MKIAA0287 protein (Fragment).
Length = 1617
Score = 34.3 bits (77), Expect = 2.6
Identities = 22/53 (41%), Positives = 26/53 (49%)
Frame = -1
Query: 693 GDEHRYPDPEAVCANEIRPEVERADPEVEAVQVRGDEVNQASVELAQTDGELE 535
G +PE A PEVE A+PEVEA + G E E A+ DGE E
Sbjct: 1417 GSNAEAAEPEVEAAE---PEVEAAEPEVEAAEPNG-EAEGPDGEAAEPDGEAE 1465
>sw|P33517|COX1_RHOSH Cytochrome c oxidase polypeptide I (EC
1.9.3.1) (Cytochrome AA3 subunit 1).
Length = 566
Score = 34.3 bits (77), Expect = 2.6
Identities = 22/76 (28%), Positives = 37/76 (48%), Gaps = 4/76 (5%)
Frame = +1
Query: 445 TVEGITAVLEGPASLLAVYAIASRKSFSHILQFAVCLGQLYGCL--VYFITAYLDGFNF- 615
TV G+T ++ AS+ Y + +H + + LG ++G +YF + G +
Sbjct: 392 TVGGVTGIVLSQASVDRYYH-DTYYVVAHF-HYVMSLGAVFGIFAGIYFWIGKMSGRQYP 449
Query: 616 -WVGPFYFWAYFIGAN 660
W G +FW F+GAN
Sbjct: 450 EWAGKLHFWMMFVGAN 465
>sptrnew|CAE50577|CAE50577 Putative membrane protein.
Length = 418
Score = 34.3 bits (77), Expect = 2.6
Identities = 19/61 (31%), Positives = 33/61 (54%), Gaps = 6/61 (9%)
Frame = -1
Query: 678 YPDPEAVCANEIRPEVERADPEVEAVQVRG------DEVNQASVELAQTDGELENVAEGL 517
+ +P A A ++ + AD E A VRG + + +A++ + QTDG++ N+A L
Sbjct: 78 HTEPVANIAQDLYSSLSLADTEASASFVRGTNDNYVEYIQKAALLVTQTDGDIANIATQL 137
Query: 516 P 514
P
Sbjct: 138 P 138
>sptr|Q8R5N0|Q8R5N0 Zinc finger protein.
Length = 1572
Score = 33.1 bits (74), Expect = 5.7
Identities = 22/53 (41%), Positives = 26/53 (49%)
Frame = -1
Query: 693 GDEHRYPDPEAVCANEIRPEVERADPEVEAVQVRGDEVNQASVELAQTDGELE 535
G +PE A PEVE A+PEVEA + G E E A+ DGE E
Sbjct: 1372 GSNAEAAEPEVEAAE---PEVEAAEPEVEAAEPIG-EAEGPDGEAAEPDGEAE 1420
>sptr|O54978|O54978 Zinc finger protein.
Length = 1571
Score = 33.1 bits (74), Expect = 5.7
Identities = 22/53 (41%), Positives = 26/53 (49%)
Frame = -1
Query: 693 GDEHRYPDPEAVCANEIRPEVERADPEVEAVQVRGDEVNQASVELAQTDGELE 535
G +PE A PEVE A+PEVEA + G E E A+ DGE E
Sbjct: 1371 GSNAEAAEPEVEAAE---PEVEAAEPEVEAAEPIG-EAEGPDGEAAEPDGEAE 1419
>sptr|Q85JB6|Q85JB6 Cytochrome c oxidase subunit 1 (EC 1.9.3.1).
Length = 487
Score = 33.1 bits (74), Expect = 5.7
Identities = 24/86 (27%), Positives = 43/86 (50%), Gaps = 6/86 (6%)
Frame = +1
Query: 445 TVEGITAVLEGPASLLAVYAIASRKSFSHILQF--AVCLGQLYGCLV--YFITAYLDGFN 612
TV G+T V+ ASL IA ++ + F + +G ++G Y+ + G+N
Sbjct: 348 TVGGVTGVILANASL----DIAFHDTYYVVAHFHYVLSMGAVFGMFAAFYYWIGKMTGYN 403
Query: 613 F--WVGPFYFWAYFIGANSFWIWIPM 684
+ ++G +FW + IG N ++ PM
Sbjct: 404 YPEFLGRLHFWTFIIGVN--LVFFPM 427
>sw|Q8IWN7|RPL1_HUMAN Retinitis pigmentosa 1-like 1 protein.
Length = 2480
Score = 33.1 bits (74), Expect = 5.7
Identities = 16/54 (29%), Positives = 27/54 (50%)
Frame = -1
Query: 678 YPDPEAVCANEIRPEVERADPEVEAVQVRGDEVNQASVELAQTDGELENVAEGL 517
+P+ E V A E E + VEA + G+ +E +T+GE + +EG+
Sbjct: 2159 HPESEDVDAQEAEGEAQPESEGVEAPEAEGEAQKAEGIEAPETEGEAQPESEGI 2212
>sptr|Q81X82|Q81X82 Phage major capsid protein, HK97 family.
Length = 346
Score = 32.7 bits (73), Expect = 7.5
Identities = 18/46 (39%), Positives = 25/46 (54%)
Frame = -1
Query: 648 EIRPEVERADPEVEAVQVRGDEVNQASVELAQTDGELENVAEGLPG 511
E+R ++E + EV A Q EV VE+ QT+ EL V + L G
Sbjct: 9 ELRAKMEAMEAEVRAEQETAQEVEVRDVEVDQTEVELRGVEQFLKG 54
Database: swall
Posted date: Feb 26, 2004 3:00 PM
Number of letters in database: 439,479,560
Number of sequences in database: 1,381,838
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 658,231,940
Number of Sequences: 1381838
Number of extensions: 13204074
Number of successful extensions: 43684
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 41161
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 43508
length of database: 439,479,560
effective HSP length: 122
effective length of database: 270,895,324
effective search space used: 47948472348
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

