BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= 8228901.3.1
(1173 letters)
Database: swall
1,381,838 sequences; 439,479,560 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
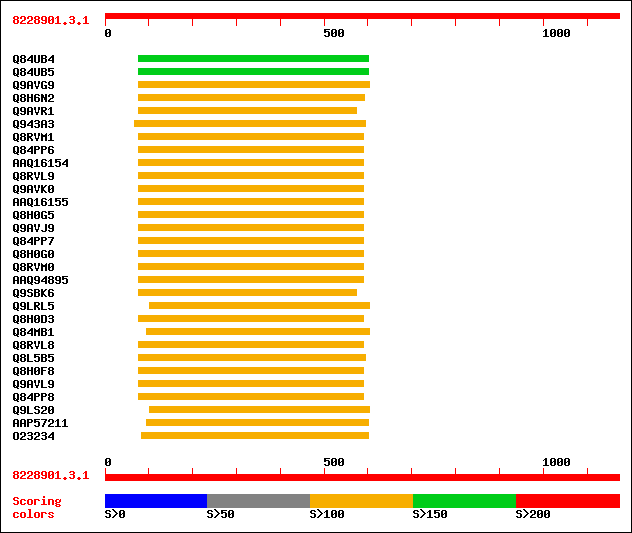
sptr|Q84UB4|Q84UB4 S-adenosyl-L-methionine:benzoic acid/salicyli... 154 3e-36
sptr|Q84UB5|Q84UB5 S-adenosyl-L-methionine:benzoic acid/salicyli... 154 3e-36
sptr|Q9AVG9|Q9AVG9 S-adenosyl-L-methionine:salicylic acid carbox... 148 2e-34
sptr|Q8H6N2|Q8H6N2 S-adenosyl-L-methionine:salicylic acid methyl... 142 8e-33
sptr|Q9AVR1|Q9AVR1 S-adenosyl-L-methionine:salicylic acid carbox... 140 3e-32
sptr|Q943A3|Q943A3 Putative S-adenosyl-L-methionine:salicylic ac... 139 8e-32
sptr|Q8RVM1|Q8RVM1 N-methyltransferase. 132 1e-29
sptr|Q84PP6|Q84PP6 Xanthosine N-methyltransferase. 132 1e-29
sptrnew|AAQ16154|AAQ16154 Putative caffeine synthase. 132 1e-29
sptr|Q8RVL9|Q8RVL9 N-methyltransferase. 132 1e-29
sptr|Q9AVK0|Q9AVK0 Theobromine synthase (N-methyltransferase) (T... 132 1e-29
sptrnew|AAQ16155|AAQ16155 Putative caffeine synthase. 132 1e-29
sptr|Q8H0G5|Q8H0G5 Theobromine synthase 1. 132 1e-29
sptr|Q9AVJ9|Q9AVJ9 7-methylxanthine N-methyltransferase. 132 1e-29
sptr|Q84PP7|Q84PP7 7-methylxanthine N-methyltransferase. 132 1e-29
sptr|Q8H0G0|Q8H0G0 Theobromine synthse 2. 132 1e-29
sptr|Q8RVM0|Q8RVM0 N-methyltransferase. 131 2e-29
sptrnew|AAQ94895|AAQ94895 Putative N-methyltransferase. 130 4e-29
sptr|Q9SBK6|Q9SBK6 Floral nectary-specific protein. 128 2e-28
sptr|Q9LRL5|Q9LRL5 S-adenosyl-L-methionine:salicylic acid carbox... 127 3e-28
sptr|Q8H0D3|Q8H0D3 Caffeine synthase 1. 127 3e-28
sptr|Q84MB1|Q84MB1 At5g38020. 127 4e-28
sptr|Q8RVL8|Q8RVL8 N-methyltransferase. 127 4e-28
sptr|Q8L5B5|Q8L5B5 Putative S-adenosyl-L-methionine:salicylic ac... 127 4e-28
sptr|Q8H0F8|Q8H0F8 Tentative caffeine synthase 4. 126 7e-28
sptr|Q9AVL9|Q9AVL9 Caffeine synthase. 126 7e-28
sptr|Q84PP8|Q84PP8 3,7-dimethylxanthine N-methyltransferase. 126 7e-28
sptr|Q9LS20|Q9LS20 S-adenosyl-L-methionine:salicylic acid carbox... 125 1e-27
sptrnew|AAP57211|AAP57211 Methyl transferase. 125 1e-27
sptr|O23234|O23234 Hypothetical protein. 125 1e-27
>sptr|Q84UB4|Q84UB4 S-adenosyl-L-methionine:benzoic acid/salicylic
acid carboxyl methyltransferase.
Length = 357
Score = 154 bits (389), Expect = 3e-36
Identities = 75/175 (42%), Positives = 112/175 (64%)
Frame = +2
Query: 77 MTIERDLHMATGNGETSYTKNSRIQEKVMFQIKPVLEEATRAAYSALLPQTMVVADLGCS 256
M + LHM GNG++SY NS +Q+KV+ KP+ E+A YS+L P+T+ +ADLGCS
Sbjct: 1 MEVVEVLHMNGGNGDSSYANNSLVQQKVILMTKPITEQAMIDLYSSLFPETLCIADLGCS 60
Query: 257 SGPNTLRFISEVIGIIARHCKEHDRRHDYPQLQFFLNDLPGNDFNNLFLLIQQFNKSIAR 436
G NT +S+++ I+ + K+H + P+ F NDLPGNDFN LF + F + + R
Sbjct: 61 LGANTFLVVSQLVKIVEKERKKHGFKS--PEFYFHFNDLPGNDFNTLFQSLGAFQEDL-R 117
Query: 437 NHKGEAAEALPPCYISGLPGSFYTRIFPSESVHLFHSLFSVHWHSQASQQLKDTQ 601
H G E+ PC+ SG+PGSFYTR+FPS+S+H +S +S+ W SQ +++ +
Sbjct: 118 KHIG---ESFGPCFFSGVPGSFYTRLFPSKSLHFVYSSYSLMWLSQVPNGIENNK 169
>sptr|Q84UB5|Q84UB5 S-adenosyl-L-methionine:benzoic acid/salicylic
acid carboxyl methyltransferase.
Length = 357
Score = 154 bits (389), Expect = 3e-36
Identities = 75/175 (42%), Positives = 112/175 (64%)
Frame = +2
Query: 77 MTIERDLHMATGNGETSYTKNSRIQEKVMFQIKPVLEEATRAAYSALLPQTMVVADLGCS 256
M + LHM GNG++SY NS +Q+KV+ KP+ E+A YS+L P+T+ +ADLGCS
Sbjct: 1 MEVVEVLHMNGGNGDSSYANNSLVQQKVILMTKPITEQAMIDLYSSLFPETLCIADLGCS 60
Query: 257 SGPNTLRFISEVIGIIARHCKEHDRRHDYPQLQFFLNDLPGNDFNNLFLLIQQFNKSIAR 436
G NT +S+++ I+ + K+H + P+ F NDLPGNDFN LF + F + + R
Sbjct: 61 LGANTFLVVSQLVKIVEKERKKHGFKS--PEFYFHFNDLPGNDFNTLFQSLGAFQEDL-R 117
Query: 437 NHKGEAAEALPPCYISGLPGSFYTRIFPSESVHLFHSLFSVHWHSQASQQLKDTQ 601
H G E+ PC+ SG+PGSFYTR+FPS+S+H +S +S+ W SQ +++ +
Sbjct: 118 KHIG---ESFGPCFFSGVPGSFYTRLFPSKSLHFVYSSYSLMWLSQVPNGIENNK 169
>sptr|Q9AVG9|Q9AVG9 S-adenosyl-L-methionine:salicylic acid carboxyl
methyltransferase.
Length = 357
Score = 148 bits (373), Expect = 2e-34
Identities = 72/176 (40%), Positives = 106/176 (60%)
Frame = +2
Query: 77 MTIERDLHMATGNGETSYTKNSRIQEKVMFQIKPVLEEATRAAYSALLPQTMVVADLGCS 256
M + LHM GNG+ SY NS +Q KV+ KP+ E+A Y + P+T+ +ADLGCS
Sbjct: 1 MKVVEVLHMNGGNGDISYANNSLVQRKVILMTKPITEQAISDLYCSFFPETLCIADLGCS 60
Query: 257 SGPNTLRFISEVIGIIARHCKEHDRRHDYPQLQFFLNDLPGNDFNNLFLLIQQFNKSIAR 436
SG NT +SE++ I+ + K H+ + F NDLPGNDFN +F + +F + + +
Sbjct: 61 SGANTFLVVSELVKIVEKERKIHNLQS--AGNLFHFNDLPGNDFNTIFQSLGKFQQDLRK 118
Query: 437 NHKGEAAEALPPCYISGLPGSFYTRIFPSESVHLFHSLFSVHWHSQASQQLKDTQK 604
+ E PC+ SG+PGSFYTR+FPSES+H HS +S+ W SQ ++ ++
Sbjct: 119 ----QIGEEFGPCFFSGVPGSFYTRLFPSESLHFVHSSYSLMWLSQVPDLIEKNKE 170
>sptr|Q8H6N2|Q8H6N2 S-adenosyl-L-methionine:salicylic acid
methyltransferase.
Length = 383
Score = 142 bits (359), Expect = 8e-33
Identities = 74/174 (42%), Positives = 109/174 (62%), Gaps = 2/174 (1%)
Frame = +2
Query: 77 MTIERDLHMATGNGETSYTKNSRIQEKVMFQIKPVLEEATRAAYSALLPQ--TMVVADLG 250
M + + LHM G G++SY NS +Q KV+ KP++EEA Y+ +LP T+ +ADLG
Sbjct: 12 MKLAQVLHMNGGLGKSSYASNSLVQRKVISITKPIIEEAMTEFYTRMLPSPHTISIADLG 71
Query: 251 CSSGPNTLRFISEVIGIIARHCKEHDRRHDYPQLQFFLNDLPGNDFNNLFLLIQQFNKSI 430
CS GPNTL +E++ II + ++ DR P+ Q LNDLPGNDFN++F + +
Sbjct: 72 CSCGPNTLLVAAELVKIIVKLRQKLDREPP-PEFQIHLNDLPGNDFNSIFRYLLPMFREE 130
Query: 431 ARNHKGEAAEALPPCYISGLPGSFYTRIFPSESVHLFHSLFSVHWHSQASQQLK 592
R G EA C++SG+PGSFY R+FP++S+H HS +S+ W S+ + +K
Sbjct: 131 LREEIGGGEEAGRRCFVSGVPGSFYGRLFPTKSLHFVHSSYSLMWLSKVPEGVK 184
>sptr|Q9AVR1|Q9AVR1 S-adenosyl-L-methionine:salicylic acid carboxyl
methyltransferase.
Length = 366
Score = 140 bits (354), Expect = 3e-32
Identities = 68/166 (40%), Positives = 101/166 (60%)
Frame = +2
Query: 77 MTIERDLHMATGNGETSYTKNSRIQEKVMFQIKPVLEEATRAAYSALLPQTMVVADLGCS 256
M + LHM G G+ SY NS +Q+KV+ KP+ EEA Y+ L P+++ +AD+GCS
Sbjct: 1 MEVVEVLHMNGGTGDASYASNSLLQKKVILLTKPITEEAITELYTRLFPKSICIADMGCS 60
Query: 257 SGPNTLRFISEVIGIIARHCKEHDRRHDYPQLQFFLNDLPGNDFNNLFLLIQQFNKSIAR 436
SGPNT +SE+I + + K H+ P+ Q LNDLP NDFN +F + F KS ++
Sbjct: 61 SGPNTFLAVSELIKNVEK--KRTSLGHESPEYQIHLNDLPSNDFNTIFRSLPSFQKSFSK 118
Query: 437 NHKGEAAEALPPCYISGLPGSFYTRIFPSESVHLFHSLFSVHWHSQ 574
+ C+ +G+PGSFY R+FP++S+H HS +S+ W S+
Sbjct: 119 ----QMGSGFGHCFFTGVPGSFYGRLFPNKSLHFVHSSYSLMWLSR 160
>sptr|Q943A3|Q943A3 Putative S-adenosyl-L-methionine:salicylic acid
carboxyl methyltransferase.
Length = 378
Score = 139 bits (350), Expect = 8e-32
Identities = 72/176 (40%), Positives = 103/176 (58%)
Frame = +2
Query: 68 AMPMTIERDLHMATGNGETSYTKNSRIQEKVMFQIKPVLEEATRAAYSALLPQTMVVADL 247
A M +E LHM G GETSY KNS +Q+K M +K ++ E+ R Y++L P+ +ADL
Sbjct: 12 APSMNVEAVLHMKEGVGETSYAKNSTLQKKSMDTVKSLVTESARDVYASLKPERFTLADL 71
Query: 248 GCSSGPNTLRFISEVIGIIARHCKEHDRRHDYPQLQFFLNDLPGNDFNNLFLLIQQFNKS 427
GCSSG N L + E++ +A C+ P+ LNDLP NDFN +F + +F
Sbjct: 72 GCSSGTNALGMVEEIVRSVAEVCRGSS---PPPEFSVLLNDLPTNDFNTIFSRLPEFTGK 128
Query: 428 IARNHKGEAAEALPPCYISGLPGSFYTRIFPSESVHLFHSLFSVHWHSQASQQLKD 595
+ + +A + P ++SG+PGSFY R+FPS++VH S S+HW SQ L D
Sbjct: 129 LKADADADAGDD-PMVFLSGVPGSFYGRLFPSKNVHFVCSFSSLHWLSQVPPGLLD 183
>sptr|Q8RVM1|Q8RVM1 N-methyltransferase.
Length = 378
Score = 132 bits (332), Expect = 1e-29
Identities = 72/174 (41%), Positives = 100/174 (57%), Gaps = 3/174 (1%)
Frame = +2
Query: 77 MTIERDLHMATGNGETSYTKNSRIQEKVMFQIKPVLEEATRAAYSALLPQT---MVVADL 247
M ++ LHM G G+TSY KNS + ++KPVLE+ R A LP + VADL
Sbjct: 1 MELQEVLHMNGGEGDTSYAKNSSYN-LALAKVKPVLEQCIRELLRANLPNINNCIKVADL 59
Query: 248 GCSSGPNTLRFISEVIGIIARHCKEHDRRHDYPQLQFFLNDLPGNDFNNLFLLIQQFNKS 427
GC+SGPNTL + +++ I + +E + P +Q FLNDL NDFN++F L+ F +
Sbjct: 60 GCASGPNTLLTVRDIVQSIDKVGQEEKNELERPTIQIFLNDLFQNDFNSVFKLLPSFYRK 119
Query: 428 IARNHKGEAAEALPPCYISGLPGSFYTRIFPSESVHLFHSLFSVHWHSQASQQL 589
+ + E + C IS +PGSFY R+FP ES+H HS +S HW SQ L
Sbjct: 120 LEK----ENGRKIGSCLISAMPGSFYGRLFPEESMHFIHSCYSFHWLSQVPSGL 169
>sptr|Q84PP6|Q84PP6 Xanthosine N-methyltransferase.
Length = 385
Score = 132 bits (331), Expect = 1e-29
Identities = 69/174 (39%), Positives = 101/174 (58%), Gaps = 3/174 (1%)
Frame = +2
Query: 77 MTIERDLHMATGNGETSYTKNSRIQEKVMFQIKPVLEEATRAAYSALLP---QTMVVADL 247
M ++ L M G G+TSY KNS + V+ ++KPVLE+ R A LP + + VADL
Sbjct: 1 MELQEVLRMNGGEGDTSYAKNSAYNQLVLAKVKPVLEQCVRELLRANLPNINKCIKVADL 60
Query: 248 GCSSGPNTLRFISEVIGIIARHCKEHDRRHDYPQLQFFLNDLPGNDFNNLFLLIQQFNKS 427
GC+SGPNTL + +++ I + +E + P +Q FLNDL NDFN++F L+ F +
Sbjct: 61 GCASGPNTLLTVRDIVQSIDKVGQEKKNELERPTIQIFLNDLFPNDFNSVFKLLPSFYRK 120
Query: 428 IARNHKGEAAEALPPCYISGLPGSFYTRIFPSESVHLFHSLFSVHWHSQASQQL 589
+ + E + C I +PGSFY+R+FP ES+H HS + + W SQ L
Sbjct: 121 LEK----ENGRKIGSCLIGAMPGSFYSRLFPEESMHFLHSCYCLQWLSQVPSGL 170
>sptrnew|AAQ16154|AAQ16154 Putative caffeine synthase.
Length = 372
Score = 132 bits (331), Expect = 1e-29
Identities = 69/174 (39%), Positives = 101/174 (58%), Gaps = 3/174 (1%)
Frame = +2
Query: 77 MTIERDLHMATGNGETSYTKNSRIQEKVMFQIKPVLEEATRAAYSALLP---QTMVVADL 247
M ++ L M G G+TSY KNS + V+ ++KPVLE+ R A LP + + VADL
Sbjct: 1 MELQEVLRMNGGEGDTSYAKNSAYNQLVLAKVKPVLEQCVRELLRANLPNINKCIKVADL 60
Query: 248 GCSSGPNTLRFISEVIGIIARHCKEHDRRHDYPQLQFFLNDLPGNDFNNLFLLIQQFNKS 427
GC+SGPNTL + +++ I + +E + P +Q FLNDL NDFN++F L+ F +
Sbjct: 61 GCASGPNTLLTVRDIVQSIDKVGQEKKNELERPTIQIFLNDLFPNDFNSVFKLLPSFYRK 120
Query: 428 IARNHKGEAAEALPPCYISGLPGSFYTRIFPSESVHLFHSLFSVHWHSQASQQL 589
+ + E + C I +PGSFY+R+FP ES+H HS + + W SQ L
Sbjct: 121 LEK----ENGRKIGSCLIGAMPGSFYSRLFPEESMHFLHSCYCLQWLSQVPSGL 170
>sptr|Q8RVL9|Q8RVL9 N-methyltransferase.
Length = 372
Score = 132 bits (331), Expect = 1e-29
Identities = 69/174 (39%), Positives = 101/174 (58%), Gaps = 3/174 (1%)
Frame = +2
Query: 77 MTIERDLHMATGNGETSYTKNSRIQEKVMFQIKPVLEEATRAAYSALLP---QTMVVADL 247
M ++ L M G G+TSY KNS + V+ ++KPVLE+ R A LP + + VADL
Sbjct: 1 MELQEVLRMNGGEGDTSYAKNSAYNQLVLAKVKPVLEQCVRELLRANLPNINKCIKVADL 60
Query: 248 GCSSGPNTLRFISEVIGIIARHCKEHDRRHDYPQLQFFLNDLPGNDFNNLFLLIQQFNKS 427
GC+SGPNTL + +++ I + +E + P +Q FLNDL NDFN++F L+ F +
Sbjct: 61 GCASGPNTLLTVRDIVQSIDKVGQEKKNELERPTIQIFLNDLFPNDFNSVFKLLPSFYRK 120
Query: 428 IARNHKGEAAEALPPCYISGLPGSFYTRIFPSESVHLFHSLFSVHWHSQASQQL 589
+ + E + C I +PGSFY+R+FP ES+H HS + + W SQ L
Sbjct: 121 LEK----ENGRKIGSCLIGAMPGSFYSRLFPEESMHFLHSCYCLQWLSQVPSGL 170
>sptr|Q9AVK0|Q9AVK0 Theobromine synthase (N-methyltransferase)
(Tentative caffeine synthase 1).
Length = 372
Score = 132 bits (331), Expect = 1e-29
Identities = 69/174 (39%), Positives = 101/174 (58%), Gaps = 3/174 (1%)
Frame = +2
Query: 77 MTIERDLHMATGNGETSYTKNSRIQEKVMFQIKPVLEEATRAAYSALLP---QTMVVADL 247
M ++ L M G G+TSY KNS + V+ ++KPVLE+ R A LP + + VADL
Sbjct: 1 MELQEVLRMNGGEGDTSYAKNSAYNQLVLAKVKPVLEQCVRELLRANLPNINKCIKVADL 60
Query: 248 GCSSGPNTLRFISEVIGIIARHCKEHDRRHDYPQLQFFLNDLPGNDFNNLFLLIQQFNKS 427
GC+SGPNTL + +++ I + +E + P +Q FLNDL NDFN++F L+ F +
Sbjct: 61 GCASGPNTLLTVRDIVQSIDKVGQEKKNELERPTIQIFLNDLFPNDFNSVFKLLPSFYRK 120
Query: 428 IARNHKGEAAEALPPCYISGLPGSFYTRIFPSESVHLFHSLFSVHWHSQASQQL 589
+ + E + C I +PGSFY+R+FP ES+H HS + + W SQ L
Sbjct: 121 LEK----ENGRKIGSCLIGAMPGSFYSRLFPEESMHFLHSCYCLQWLSQVPSGL 170
>sptrnew|AAQ16155|AAQ16155 Putative caffeine synthase.
Length = 378
Score = 132 bits (331), Expect = 1e-29
Identities = 71/174 (40%), Positives = 101/174 (58%), Gaps = 3/174 (1%)
Frame = +2
Query: 77 MTIERDLHMATGNGETSYTKNSRIQEKVMFQIKPVLEEATRAAYSALLP---QTMVVADL 247
M ++ LHM G G+TSY KN+ + ++KP LE+ R A LP + + VADL
Sbjct: 1 MELQEVLHMNEGEGDTSYAKNASYN-LALAKVKPFLEQCIRELLRANLPNINKCIKVADL 59
Query: 248 GCSSGPNTLRFISEVIGIIARHCKEHDRRHDYPQLQFFLNDLPGNDFNNLFLLIQQFNKS 427
GC+SGPNTL + +++ I + +E + P +Q FLNDL NDFN++F L+ F +
Sbjct: 60 GCASGPNTLLTVRDIVQSIDKVGQEEKNELERPTIQIFLNDLFQNDFNSVFKLLPSFYRK 119
Query: 428 IARNHKGEAAEALPPCYISGLPGSFYTRIFPSESVHLFHSLFSVHWHSQASQQL 589
+ + E + C IS +PGSFY R+FP ES+H HS +SVHW SQ L
Sbjct: 120 LEK----ENGRKIGSCLISAMPGSFYGRLFPEESMHFLHSCYSVHWLSQVPSGL 169
>sptr|Q8H0G5|Q8H0G5 Theobromine synthase 1.
Length = 378
Score = 132 bits (331), Expect = 1e-29
Identities = 71/174 (40%), Positives = 101/174 (58%), Gaps = 3/174 (1%)
Frame = +2
Query: 77 MTIERDLHMATGNGETSYTKNSRIQEKVMFQIKPVLEEATRAAYSALLP---QTMVVADL 247
M ++ LHM G G+TSY KN+ + ++KP LE+ R A LP + + VADL
Sbjct: 1 MELQEVLHMNEGEGDTSYAKNASYN-LALAKVKPFLEQCIRELLRANLPNINKCIKVADL 59
Query: 248 GCSSGPNTLRFISEVIGIIARHCKEHDRRHDYPQLQFFLNDLPGNDFNNLFLLIQQFNKS 427
GC+SGPNTL + +++ I + +E + P +Q FLNDL NDFN++F L+ F +
Sbjct: 60 GCASGPNTLLTVRDIVQSIDKVGQEEKNELERPTIQIFLNDLFQNDFNSVFKLLPSFYRK 119
Query: 428 IARNHKGEAAEALPPCYISGLPGSFYTRIFPSESVHLFHSLFSVHWHSQASQQL 589
+ + E + C IS +PGSFY R+FP ES+H HS +SVHW SQ L
Sbjct: 120 LEK----ENGRKIGSCLISAMPGSFYGRLFPEESMHFLHSCYSVHWLSQVPSGL 169
>sptr|Q9AVJ9|Q9AVJ9 7-methylxanthine N-methyltransferase.
Length = 378
Score = 132 bits (331), Expect = 1e-29
Identities = 71/174 (40%), Positives = 101/174 (58%), Gaps = 3/174 (1%)
Frame = +2
Query: 77 MTIERDLHMATGNGETSYTKNSRIQEKVMFQIKPVLEEATRAAYSALLP---QTMVVADL 247
M ++ LHM G G+TSY KN+ + ++KP LE+ R A LP + + VADL
Sbjct: 1 MELQEVLHMNEGEGDTSYAKNASYN-LALAKVKPFLEQCIRELLRANLPNINKCIKVADL 59
Query: 248 GCSSGPNTLRFISEVIGIIARHCKEHDRRHDYPQLQFFLNDLPGNDFNNLFLLIQQFNKS 427
GC+SGPNTL + +++ I + +E + P +Q FLNDL NDFN++F L+ F +
Sbjct: 60 GCASGPNTLLTVRDIVQSIDKVGQEEKNELERPTIQIFLNDLFQNDFNSVFKLLPSFYRK 119
Query: 428 IARNHKGEAAEALPPCYISGLPGSFYTRIFPSESVHLFHSLFSVHWHSQASQQL 589
+ + E + C IS +PGSFY R+FP ES+H HS +SVHW SQ L
Sbjct: 120 LEK----ENGRKIGSCLISAMPGSFYGRLFPEESMHFLHSCYSVHWLSQVPSGL 169
>sptr|Q84PP7|Q84PP7 7-methylxanthine N-methyltransferase.
Length = 384
Score = 132 bits (331), Expect = 1e-29
Identities = 71/174 (40%), Positives = 101/174 (58%), Gaps = 3/174 (1%)
Frame = +2
Query: 77 MTIERDLHMATGNGETSYTKNSRIQEKVMFQIKPVLEEATRAAYSALLP---QTMVVADL 247
M ++ LHM G G+TSY KN+ + ++KP LE+ R A LP + + VADL
Sbjct: 1 MELQEVLHMNEGEGDTSYAKNASYN-LALAKVKPFLEQCIRELLRANLPNINKCIKVADL 59
Query: 248 GCSSGPNTLRFISEVIGIIARHCKEHDRRHDYPQLQFFLNDLPGNDFNNLFLLIQQFNKS 427
GC+SGPNTL + +++ I + +E + P +Q FLNDL NDFN++F L+ F +
Sbjct: 60 GCASGPNTLLTVRDIVQSIDKVGQEEKNELERPTIQIFLNDLFQNDFNSVFKLLPSFYRK 119
Query: 428 IARNHKGEAAEALPPCYISGLPGSFYTRIFPSESVHLFHSLFSVHWHSQASQQL 589
+ + E + C IS +PGSFY R+FP ES+H HS +SVHW SQ L
Sbjct: 120 LEK----ENGRKIGSCLISAMPGSFYGRLFPEESMHFLHSCYSVHWLSQVPSGL 169
>sptr|Q8H0G0|Q8H0G0 Theobromine synthse 2.
Length = 384
Score = 132 bits (331), Expect = 1e-29
Identities = 72/174 (41%), Positives = 100/174 (57%), Gaps = 3/174 (1%)
Frame = +2
Query: 77 MTIERDLHMATGNGETSYTKNSRIQEKVMFQIKPVLEEATRAAYSALLPQT---MVVADL 247
M ++ LHM G G+TSY KNS + ++KPVLE+ R A LP + VADL
Sbjct: 1 MELQAVLHMNGGEGDTSYAKNSSYN-LALAKVKPVLEQCIRELLRANLPNINNCIKVADL 59
Query: 248 GCSSGPNTLRFISEVIGIIARHCKEHDRRHDYPQLQFFLNDLPGNDFNNLFLLIQQFNKS 427
GC+SGPNTL + +++ I + +E + P +Q FLNDL NDFN++F L+ F +
Sbjct: 60 GCASGPNTLLTVRDIVQSIDKVGQEEKNELERPTIQIFLNDLFQNDFNSVFKLLPSFYRK 119
Query: 428 IARNHKGEAAEALPPCYISGLPGSFYTRIFPSESVHLFHSLFSVHWHSQASQQL 589
+ + E + C IS +PGSFY R+FP ES+H HS +S HW SQ L
Sbjct: 120 LEK----ENGRKIGSCLISAMPGSFYGRLFPEESMHFIHSCYSFHWLSQVPSGL 169
>sptr|Q8RVM0|Q8RVM0 N-methyltransferase.
Length = 371
Score = 131 bits (330), Expect = 2e-29
Identities = 71/174 (40%), Positives = 100/174 (57%), Gaps = 3/174 (1%)
Frame = +2
Query: 77 MTIERDLHMATGNGETSYTKNSRIQEKVMFQIKPVLEEATRAAYSALLPQT---MVVADL 247
M ++ LHM G G+TSY KN+ + ++KP LE+ R A LP + VADL
Sbjct: 1 MELQEVLHMNEGEGDTSYAKNASYN-LALAKVKPFLEQCIRELLRANLPNINKYIKVADL 59
Query: 248 GCSSGPNTLRFISEVIGIIARHCKEHDRRHDYPQLQFFLNDLPGNDFNNLFLLIQQFNKS 427
GC+SGPNTL + +++ I + +E + P +Q FLNDL NDFN++F L+ F +
Sbjct: 60 GCASGPNTLLTVRDIVQSIDKVGQEEKNELERPTIQIFLNDLFQNDFNSVFKLLPSFYRK 119
Query: 428 IARNHKGEAAEALPPCYISGLPGSFYTRIFPSESVHLFHSLFSVHWHSQASQQL 589
+ + E + C IS +PGSFY R+FP ES+H HS +SVHW SQ L
Sbjct: 120 LEK----ENGRKIGSCLISAMPGSFYGRLFPEESMHFLHSCYSVHWLSQVPSGL 169
>sptrnew|AAQ94895|AAQ94895 Putative N-methyltransferase.
Length = 384
Score = 130 bits (327), Expect = 4e-29
Identities = 71/174 (40%), Positives = 100/174 (57%), Gaps = 3/174 (1%)
Frame = +2
Query: 77 MTIERDLHMATGNGETSYTKNSRIQEKVMFQIKPVLEEATRAAYSALLPQT---MVVADL 247
M ++ LHM G G+TSY KNS + ++KPVLE+ R A LP + VADL
Sbjct: 1 MELQEVLHMNGGEGDTSYAKNSSYN-LALAKVKPVLEQCIRELLRANLPNINNCIKVADL 59
Query: 248 GCSSGPNTLRFISEVIGIIARHCKEHDRRHDYPQLQFFLNDLPGNDFNNLFLLIQQFNKS 427
GC+SGPNTL + +++ I + E + P +Q FLNDL NDFN++F L+ F +
Sbjct: 60 GCASGPNTLLTVRDIVQSIDKVGLEEKNELERPTVQIFLNDLFQNDFNSVFKLLPSFYRK 119
Query: 428 IARNHKGEAAEALPPCYISGLPGSFYTRIFPSESVHLFHSLFSVHWHSQASQQL 589
+ + E + C IS +PGSF+ R+FP ES+H HS +S+HW SQ L
Sbjct: 120 LEK----ENGRKIGSCLISAMPGSFHGRLFPEESMHFLHSCYSIHWLSQVPSGL 169
>sptr|Q9SBK6|Q9SBK6 Floral nectary-specific protein.
Length = 392
Score = 128 bits (321), Expect = 2e-28
Identities = 71/174 (40%), Positives = 98/174 (56%), Gaps = 8/174 (4%)
Frame = +2
Query: 77 MTIERDLHMATGNGETSYTKNSRIQEKVMFQIKPVLEEATRAAY---SALLPQTMVVADL 247
M + R LHM GNGETSY KNS +Q ++ + V++EA + S +L + +ADL
Sbjct: 1 MEVMRILHMNKGNGETSYAKNSIVQSNIISLGRRVMDEALKKLMIRNSEIL--SFGIADL 58
Query: 248 GCSSGPNTLRFISEVIGIIARHCKEHDRRHDYPQLQFFLNDLPGNDFNNLFLLIQQFNKS 427
GCSSGPN+L IS ++ I C HD P+L LNDLP NDFN +F + +F
Sbjct: 59 GCSSGPNSLLSISNIVETIQNLC--HDLDRPVPELSLSLNDLPSNDFNYIFASLPEFYDR 116
Query: 428 IARNHKGEAAEALP-----PCYISGLPGSFYTRIFPSESVHLFHSLFSVHWHSQ 574
+ + + PC++S +PGSFY R+FP S+H HS S+HW SQ
Sbjct: 117 VKKRDNNYESLGFEHGSGGPCFVSAVPGSFYGRLFPRRSLHFVHSSSSLHWLSQ 170
>sptr|Q9LRL5|Q9LRL5 S-adenosyl-L-methionine:salicylic acid carboxyl
methyltransferase-like protein, floral nectary-specific
protein-like.
Length = 368
Score = 127 bits (320), Expect = 3e-28
Identities = 68/169 (40%), Positives = 96/169 (56%), Gaps = 1/169 (0%)
Frame = +2
Query: 101 MATGNGETSYTKNSRIQEKVMFQIKP-VLEEATRAAYSALLPQTMVVADLGCSSGPNTLR 277
M G+GE SY NS Q+ + KP V++ P + VADLGCSSG NT
Sbjct: 6 MKGGDGEHSYANNSEAQKSITSDAKPEVMKSVNEMIVKMDFPGCIKVADLGCSSGENTFL 65
Query: 278 FISEVIGIIARHCKEHDRRHDYPQLQFFLNDLPGNDFNNLFLLIQQFNKSIARNHKGEAA 457
+SE++ I +++ + + P++ LNDLP NDFN F LI F++ + N KG
Sbjct: 66 VMSEIVNTIITTYQQNGQ--NLPEIDCCLNDLPENDFNTTFKLIPSFHEKLKMNVKGN-- 121
Query: 458 EALPPCYISGLPGSFYTRIFPSESVHLFHSLFSVHWHSQASQQLKDTQK 604
CY+SG PGSFYTR+FPS+S+H HS F +HW S+ L++ +K
Sbjct: 122 -----CYVSGCPGSFYTRLFPSKSLHFVHSSFCLHWLSKVPDGLEENKK 165
>sptr|Q8H0D3|Q8H0D3 Caffeine synthase 1.
Length = 384
Score = 127 bits (319), Expect = 3e-28
Identities = 69/174 (39%), Positives = 98/174 (56%), Gaps = 3/174 (1%)
Frame = +2
Query: 77 MTIERDLHMATGNGETSYTKNSRIQEKVMFQIKPVLEEATRAAYSALLP---QTMVVADL 247
M ++ LHM G G+TSY KNS + ++KPVLE+ + A LP + V DL
Sbjct: 1 MELQEVLHMNGGEGDTSYAKNSSYN-LFLIRVKPVLEQCIQELLRANLPNINKCFKVGDL 59
Query: 248 GCSSGPNTLRFISEVIGIIARHCKEHDRRHDYPQLQFFLNDLPGNDFNNLFLLIQQFNKS 427
GC+SGPNT + +++ I + +E + P +Q FLNDL NDFN++F L+ F
Sbjct: 60 GCASGPNTFSTVRDIVQSIDKVGQEKKNELERPTIQIFLNDLFQNDFNSVFKLLPSFY-- 117
Query: 428 IARNHKGEAAEALPPCYISGLPGSFYTRIFPSESVHLFHSLFSVHWHSQASQQL 589
RN + E + C I +PGSFY+R+FP ES+H HS + +HW SQ L
Sbjct: 118 --RNLEKENGRKIGSCLIGAMPGSFYSRLFPEESMHFLHSCYCLHWLSQVPSGL 169
>sptr|Q84MB1|Q84MB1 At5g38020.
Length = 368
Score = 127 bits (318), Expect = 4e-28
Identities = 69/171 (40%), Positives = 100/171 (58%), Gaps = 1/171 (0%)
Frame = +2
Query: 95 LHMATGNGETSYTKNSRIQEKVMFQIKPVLEEATRAAYSAL-LPQTMVVADLGCSSGPNT 271
L M G+GE SY NS Q+++ KPV+ E + P + VADLGCSSG NT
Sbjct: 4 LSMKGGDGEHSYANNSEGQKRLASDAKPVVVETVKEMIVKTDFPGCIKVADLGCSSGENT 63
Query: 272 LRFISEVIGIIARHCKEHDRRHDYPQLQFFLNDLPGNDFNNLFLLIQQFNKSIARNHKGE 451
L +SE++ I ++ + + P++ LNDLP NDFN F L+ F+K + + KG+
Sbjct: 64 LLVMSEIVNTIITSYQQKGK--NLPEINCCLNDLPDNDFNTTFKLVPAFHKLLKMDVKGK 121
Query: 452 AAEALPPCYISGLPGSFYTRIFPSESVHLFHSLFSVHWHSQASQQLKDTQK 604
C+ISG+PGSFY+R+FPS+S+H HS +HW S+ L+D +K
Sbjct: 122 -------CFISGVPGSFYSRLFPSKSLHFVHSSLCLHWLSKVPDGLEDNKK 165
>sptr|Q8RVL8|Q8RVL8 N-methyltransferase.
Length = 386
Score = 127 bits (318), Expect = 4e-28
Identities = 70/174 (40%), Positives = 98/174 (56%), Gaps = 3/174 (1%)
Frame = +2
Query: 77 MTIERDLHMATGNGETSYTKNSRIQEKVMFQIKPVLEEATRAAYSALLPQT---MVVADL 247
M ++ L M G G+TSY KNS + ++KPVLE+ R A LP + VADL
Sbjct: 1 MELQEVLRMNGGEGDTSYAKNSSYN-LALAKVKPVLEQCIRELLRANLPNINNCIKVADL 59
Query: 248 GCSSGPNTLRFISEVIGIIARHCKEHDRRHDYPQLQFFLNDLPGNDFNNLFLLIQQFNKS 427
GC+SGPNTL + +++ I + E + P +Q FLNDL NDFN++F L+ F +
Sbjct: 60 GCASGPNTLLTVRDIVQSIDKLGLEEKNELERPTIQIFLNDLFQNDFNSVFKLLPSFYRK 119
Query: 428 IARNHKGEAAEALPPCYISGLPGSFYTRIFPSESVHLFHSLFSVHWHSQASQQL 589
+ + E + C IS +PGSFY R+FP ES+H HS + +HW SQ L
Sbjct: 120 LEK----ENGRKIGSCLISAMPGSFYGRLFPEESMHFLHSCYCLHWLSQVPSGL 169
>sptr|Q8L5B5|Q8L5B5 Putative S-adenosyl-L-methionine:salicylic acid
carboxyl methyltransferase.
Length = 375
Score = 127 bits (318), Expect = 4e-28
Identities = 71/173 (41%), Positives = 95/173 (54%)
Frame = +2
Query: 77 MTIERDLHMATGNGETSYTKNSRIQEKVMFQIKPVLEEATRAAYSALLPQTMVVADLGCS 256
M +E LHM G GETSY +NSR M +K ++ + Y + +P+ VADLGCS
Sbjct: 15 MNVETVLHMKEGLGETSYAQNSR----GMDTLKSLITNSAADVYLSQMPERFAVADLGCS 70
Query: 257 SGPNTLRFISEVIGIIARHCKEHDRRHDYPQLQFFLNDLPGNDFNNLFLLIQQFNKSIAR 436
SGPN L ++IG I R C R P+ LNDLP NDFN +F + +F +
Sbjct: 71 SGPNALCLAEDIIGSIGRICCRSSRPP--PEFSVLLNDLPTNDFNTIFFSLPEFTDRLKA 128
Query: 437 NHKGEAAEALPPCYISGLPGSFYTRIFPSESVHLFHSLFSVHWHSQASQQLKD 595
K + P ++SG+PGSFY R+FP++SVH S S+HW SQ L D
Sbjct: 129 AAKSDEW-GRPMVFLSGVPGSFYGRLFPAKSVHFVCSCSSLHWLSQVPSGLLD 180
>sptr|Q8H0F8|Q8H0F8 Tentative caffeine synthase 4.
Length = 385
Score = 126 bits (316), Expect = 7e-28
Identities = 68/174 (39%), Positives = 101/174 (58%), Gaps = 3/174 (1%)
Frame = +2
Query: 77 MTIERDLHMATGNGETSYTKNSRIQEKVMFQIKPVLEEATRAAYSALLP---QTMVVADL 247
M ++ LHM G GE SY KNS + V+ ++KPVLE+ R A LP + + VADL
Sbjct: 1 MELQEVLHMNGGEGEASYAKNSSFNQLVLAKVKPVLEQCVRELLRANLPNINKCIKVADL 60
Query: 248 GCSSGPNTLRFISEVIGIIARHCKEHDRRHDYPQLQFFLNDLPGNDFNNLFLLIQQFNKS 427
GC+SGPNTL + + + I + +E + P +Q FL DL NDFN++F+L+ F +
Sbjct: 61 GCASGPNTLLTVWDTVQSIDKVRQEMKNELERPTIQVFLTDLFQNDFNSVFMLLPSFYRK 120
Query: 428 IARNHKGEAAEALPPCYISGLPGSFYTRIFPSESVHLFHSLFSVHWHSQASQQL 589
+ + E + C I+ +PGSF+ R+FP ES+H HS +S+ + SQ L
Sbjct: 121 LEK----ENGRKIGSCLIAAMPGSFHGRLFPEESMHFLHSSYSLQFLSQVPSGL 170
>sptr|Q9AVL9|Q9AVL9 Caffeine synthase.
Length = 385
Score = 126 bits (316), Expect = 7e-28
Identities = 68/174 (39%), Positives = 101/174 (58%), Gaps = 3/174 (1%)
Frame = +2
Query: 77 MTIERDLHMATGNGETSYTKNSRIQEKVMFQIKPVLEEATRAAYSALLP---QTMVVADL 247
M ++ LHM G GE SY KNS + V+ ++KPVLE+ R A LP + + VADL
Sbjct: 1 MELQEVLHMNGGEGEASYAKNSSFNQLVLAKVKPVLEQCVRELLRANLPNINKCIKVADL 60
Query: 248 GCSSGPNTLRFISEVIGIIARHCKEHDRRHDYPQLQFFLNDLPGNDFNNLFLLIQQFNKS 427
GC+SGPNTL + + + I + +E + P +Q FL DL NDFN++F+L+ F +
Sbjct: 61 GCASGPNTLLTVWDTVQSIDKVKQEMKNELERPTIQVFLTDLFQNDFNSVFMLLPSFYRK 120
Query: 428 IARNHKGEAAEALPPCYISGLPGSFYTRIFPSESVHLFHSLFSVHWHSQASQQL 589
+ + E + C I+ +PGSF+ R+FP ES+H HS +S+ + SQ L
Sbjct: 121 LEK----ENGRKIGSCLIAAMPGSFHGRLFPEESMHFLHSSYSLQFLSQVPSGL 170
>sptr|Q84PP8|Q84PP8 3,7-dimethylxanthine N-methyltransferase.
Length = 384
Score = 126 bits (316), Expect = 7e-28
Identities = 67/174 (38%), Positives = 99/174 (56%), Gaps = 3/174 (1%)
Frame = +2
Query: 77 MTIERDLHMATGNGETSYTKNSRIQEKVMFQIKPVLEEATRAAYSALLP---QTMVVADL 247
M ++ LHM G G+TSY KNS + ++KP+LE+ + A LP + + VADL
Sbjct: 1 MELQEVLHMNGGEGDTSYAKNS-FYNLFLIRVKPILEQCIQELLRANLPNINKCIKVADL 59
Query: 248 GCSSGPNTLRFISEVIGIIARHCKEHDRRHDYPQLQFFLNDLPGNDFNNLFLLIQQFNKS 427
GC+SGPNTL + +++ I + +E + P +Q FLNDL NDFN++F + F +
Sbjct: 60 GCASGPNTLLTVRDIVQSIDKVGQEKKNELERPTIQIFLNDLFQNDFNSVFKSLPSFYRK 119
Query: 428 IARNHKGEAAEALPPCYISGLPGSFYTRIFPSESVHLFHSLFSVHWHSQASQQL 589
+ + E + C I +PGSFY R+FP ES+H HS + +HW SQ L
Sbjct: 120 LEK----ENGRKIGSCLIGAMPGSFYGRLFPEESMHFLHSCYCLHWLSQVPSGL 169
>sptr|Q9LS20|Q9LS20 S-adenosyl-L-methionine:salicylic acid carboxyl
methyltransferase- like.
Length = 363
Score = 125 bits (315), Expect = 1e-27
Identities = 68/169 (40%), Positives = 99/169 (58%), Gaps = 1/169 (0%)
Frame = +2
Query: 101 MATGNGETSYTKNSRIQEKVMFQIKPVLEEATRAAYSAL-LPQTMVVADLGCSSGPNTLR 277
M G+GE SY NS Q+++ KPV+ E + P + VADLGCSSG NTL
Sbjct: 1 MKGGDGEHSYANNSEGQKRLASDAKPVVVETVKEMIVKTDFPGCIKVADLGCSSGENTLL 60
Query: 278 FISEVIGIIARHCKEHDRRHDYPQLQFFLNDLPGNDFNNLFLLIQQFNKSIARNHKGEAA 457
+SE++ I ++ + + P++ LNDLP NDFN F L+ F+K + + KG+
Sbjct: 61 VMSEIVNTIITSYQQKGK--NLPEINCCLNDLPDNDFNTTFKLVPAFHKLLKMDVKGK-- 116
Query: 458 EALPPCYISGLPGSFYTRIFPSESVHLFHSLFSVHWHSQASQQLKDTQK 604
C+ISG+PGSFY+R+FPS+S+H HS +HW S+ L+D +K
Sbjct: 117 -----CFISGVPGSFYSRLFPSKSLHFVHSSLCLHWLSKVPDGLEDNKK 160
>sptrnew|AAP57211|AAP57211 Methyl transferase.
Length = 380
Score = 125 bits (315), Expect = 1e-27
Identities = 67/170 (39%), Positives = 100/170 (58%), Gaps = 1/170 (0%)
Frame = +2
Query: 95 LHMATGNGETSYTKNSRIQEKVMFQIKPVLEEATRAAYSAL-LPQTMVVADLGCSSGPNT 271
L M+ G+G SY+ NSR+Q KV+ KPVL + T +L P + VA+LGCSSG NT
Sbjct: 29 LCMSGGDGTNSYSANSRLQRKVLTMAKPVLVKTTEEMMMSLDFPTYIKVAELGCSSGQNT 88
Query: 272 LRFISEVIGIIARHCKEHDRRHDYPQLQFFLNDLPGNDFNNLFLLIQQFNKSIARNHKGE 451
ISE+I I+ C+ ++ P++ LNDLP NDFN F + FNK + K
Sbjct: 89 FLAISEIINTISVLCQHVNKNP--PEIDCCLNDLPENDFNTTFKFVPFFNKELMITSKAS 146
Query: 452 AAEALPPCYISGLPGSFYTRIFPSESVHLFHSLFSVHWHSQASQQLKDTQ 601
C++ G PGSFY+R+F S+H+ HS +++HW S+ ++L++ +
Sbjct: 147 -------CFVYGAPGSFYSRLFSRNSLHIIHSSYALHWLSKVPEKLENNK 189
>sptr|O23234|O23234 Hypothetical protein.
Length = 371
Score = 125 bits (315), Expect = 1e-27
Identities = 65/174 (37%), Positives = 98/174 (56%), Gaps = 1/174 (0%)
Frame = +2
Query: 83 IERDLHMATGNGETSYTKNSRIQEKVMFQIKPVLEEATRAAYSALLPQTMVVADLGCSSG 262
+ER+ +M G+G+TSY +NS +Q+K K + E + Y P+++ +ADLGCSSG
Sbjct: 6 MEREFYMTGGDGKTSYARNSSLQKKASDTAKHITLETLQQLYKETRPKSLGIADLGCSSG 65
Query: 263 PNTLRFISEVIGIIARHCKEHDRRHDYPQLQFFLNDLPGNDFNNLFLLIQQFNKSIAR-N 439
PNTL I++ I + P+ FLNDLPGNDFN +F + F+ + R N
Sbjct: 66 PNTLSTITDFIKTVQVAHHREIPIQPLPEFSIFLNDLPGNDFNFIFKSLPDFHIELKRDN 125
Query: 440 HKGEAAEALPPCYISGLPGSFYTRIFPSESVHLFHSLFSVHWHSQASQQLKDTQ 601
+ G+ P +I+ PGSFY R+FP ++H ++ S+HW S+ L D Q
Sbjct: 126 NNGDC----PSVFIAAYPGSFYGRLFPENTIHFVYASHSLHWLSKVPTALYDEQ 175
Database: swall
Posted date: Feb 26, 2004 3:00 PM
Number of letters in database: 439,479,560
Number of sequences in database: 1,381,838
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,013,521,764
Number of Sequences: 1381838
Number of extensions: 22743556
Number of successful extensions: 50400
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 47035
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 50102
length of database: 439,479,560
effective HSP length: 125
effective length of database: 266,749,810
effective search space used: 70688699650
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

