BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= 5705280.3.1
(1124 letters)
Database: swall
1,381,838 sequences; 439,479,560 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
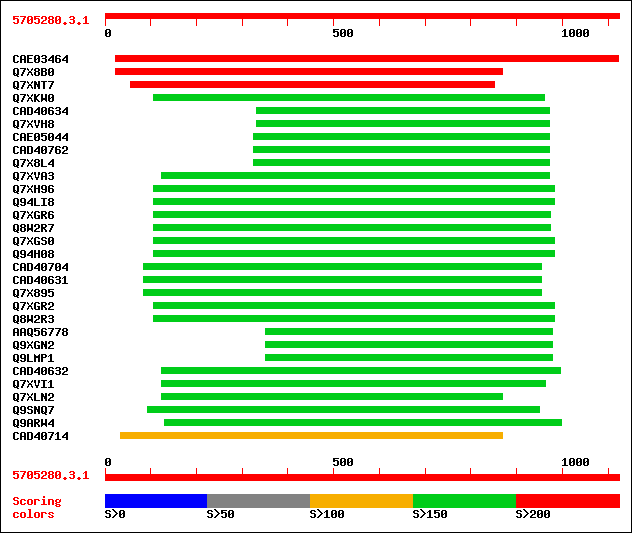
sptrnew|CAE03464|CAE03464 OSJNBa0083N12.1 protein. 455 e-127
sptr|Q7X8B0|Q7X8B0 OSJNBa0083N12.1 protein (OSJNBa0093F12.22 pro... 354 1e-96
sptr|Q7XNT7|Q7XNT7 OSJNBa0093F12.21 protein. 206 7e-52
sptr|Q7XKW0|Q7XKW0 OSJNBa0073E02.9 protein. 181 2e-44
sptrnew|CAD40634|CAD40634 OSJNBa0016N04.10 protein. 180 3e-44
sptr|Q7XVH8|Q7XVH8 OSJNBa0016N04.12 protein. 180 3e-44
sptrnew|CAE05044|CAE05044 OSJNBa0049H08.5 protein. 177 2e-43
sptrnew|CAD40762|CAD40762 OSJNBa0081G05.15 protein. 177 2e-43
sptr|Q7X8L4|Q7X8L4 OSJNBa0081G05.22 protein (OSJNBa0049H08.6 pro... 177 2e-43
sptr|Q7XVA3|Q7XVA3 OSJNBa0081G05.12 protein. 177 3e-43
sptr|Q7XH96|Q7XH96 Putative wall-associated protein kinase. 170 3e-41
sptr|Q94LI8|Q94LI8 Putative wall-associated protein kinase. 170 3e-41
sptr|Q7XGR6|Q7XGR6 Putative wall-associated protein kinase. 166 8e-40
sptr|Q8W2R7|Q8W2R7 Putative wall-associated protein kinase. 166 8e-40
sptr|Q7XGS0|Q7XGS0 Putative wall-associated protein kinase. 166 8e-40
sptr|Q94H08|Q94H08 Putative wall-associated protein kinase. 166 8e-40
sptrnew|CAD40704|CAD40704 OSJNBb0042I07.1 protein. 164 3e-39
sptrnew|CAD40631|CAD40631 OSJNBa0016N04.13 protein. 164 3e-39
sptr|Q7X895|Q7X895 OSJNBa0016N04.17 protein (OSJNBb0042I07.2 pro... 164 3e-39
sptr|Q7XGR2|Q7XGR2 Putative wall-associated protein kinase. 160 3e-38
sptr|Q8W2R3|Q8W2R3 Putative wall-associated protein kinase. 160 3e-38
sptrnew|AAQ56778|AAQ56778 At1g21270. 160 4e-38
sptr|Q9XGN2|Q9XGN2 Wall-associated kinase 2 (Hypothetical protein). 160 4e-38
sptr|Q9LMP1|Q9LMP1 F16F4.5 protein (Wall-associated kinase 2). 160 4e-38
sptrnew|CAD40632|CAD40632 OSJNBa0016N04.12 protein. 157 2e-37
sptr|Q7XVI1|Q7XVI1 OSJNBa0016N04.15 protein. 154 2e-36
sptr|Q7XLN2|Q7XLN2 OSJNBa0049H08.9 protein. 154 2e-36
sptr|Q9SNQ7|Q9SNQ7 Similar to wak1 gene. 154 2e-36
sptr|Q9ARW4|Q9ARW4 Putative wall-associated kinase 2. 152 7e-36
sptrnew|CAD40714|CAD40714 OSJNBb0042I07.11 protein. 149 1e-34
>sptrnew|CAE03464|CAE03464 OSJNBa0083N12.1 protein.
Length = 712
Score = 455 bits (1170), Expect = e-127
Identities = 223/370 (60%), Positives = 265/370 (71%), Gaps = 3/370 (0%)
Frame = +1
Query: 22 VLLRLWHIPFFL-TCFSPSFVHPMEPGTYRCSNVSIPIPYPFGVAGKSPSLSRGFKVVCG 198
++L L +P L TC S+VH ME T +CSN IPIPYPFG+ G +P+ ++GF++ C
Sbjct: 4 MMLYLLLLPLLLLTCIFASYVHSMESSTSKCSN--IPIPYPFGILGGNPAPAQGFEITCA 61
Query: 199 HSGPLLSIGNSMXXXXXXXXXXXXXXXXASASSQQCIGGTANSSVNLEGTVFTFSDTRNK 378
SGP++ I N M ASA+SQQC NSS +LEGT FTFSDTRNK
Sbjct: 62 SSGPMVRINNIMFGILNISLLDGFVSILASATSQQC---KRNSSFSLEGTNFTFSDTRNK 118
Query: 379 FTALGCNVVAMLLNGSTGYSGGCASFCSSGNNIIDGSCSGVACCQAPVPKGLKKLDLEFS 558
FTALGC++VAMLLNGS+GYSGGCASFCS+ +NIIDG CSGVACCQAPVPKGLKKL+LEF+
Sbjct: 119 FTALGCDMVAMLLNGSSGYSGGCASFCSTKSNIIDGMCSGVACCQAPVPKGLKKLELEFT 178
Query: 559 NIN--ITNSGIDYDYWPCGQAFVVEQDSYVFSALDLNNTNDTKPQYRHVVLEWSVNGSSC 732
NI ++ + CG+AF+VEQ+SYVFS++DL+NTN PQYR VVLEWS++G C
Sbjct: 179 NITGQLSRPKEVNNTPTCGEAFIVEQNSYVFSSVDLSNTNRNNPQYRPVVLEWSIDGGYC 238
Query: 733 EEAKLSGSYACAENAYCYNSSNGIGYRCNCSSGFEGNPYLQGPNGCTDINECITRNPCTN 912
EEA SYAC EN+YCYNSSNGIGYRCNCS GF+GNPYLQGP+GC DI+EC + PCT+
Sbjct: 239 EEANRFMSYACKENSYCYNSSNGIGYRCNCSLGFQGNPYLQGPDGCQDIDECTIKRPCTH 298
Query: 913 RCSNTIGGFQCTCPAGMSGDGLKEGSGCNGVSTXXXXXXXXXXXXXXXXXXXFWTHWLGK 1092
+C NT G F C CPAGM GDGLKEGSGCNG+ T FWTHWL K
Sbjct: 299 KCINTKGSFYCMCPAGMRGDGLKEGSGCNGIGTLLIGIVTGLALLLLLLVLIFWTHWLVK 358
Query: 1093 KRKLAKTRQR 1122
KRKLAK RQR
Sbjct: 359 KRKLAKIRQR 368
>sptr|Q7X8B0|Q7X8B0 OSJNBa0083N12.1 protein (OSJNBa0093F12.22
protein).
Length = 661
Score = 354 bits (909), Expect = 1e-96
Identities = 176/286 (61%), Positives = 213/286 (74%), Gaps = 3/286 (1%)
Frame = +1
Query: 22 VLLRLWHIPFFL-TCFSPSFVHPMEPGTYRCSNVSIPIPYPFGVAGKSPSLSRGFKVVCG 198
++L L +P L TC S+VH ME T +CSN IPIPYPFG+ G +P+ ++GF++ C
Sbjct: 4 MMLYLLLLPLLLLTCIFASYVHSMESSTSKCSN--IPIPYPFGILGGNPAPAQGFEITCA 61
Query: 199 HSGPLLSIGNSMXXXXXXXXXXXXXXXXASASSQQCIGGTANSSVNLEGTVFTFSDTRNK 378
SGP++ I N M ASA+SQQC NSS +LEGT FTFSDTRNK
Sbjct: 62 SSGPMVRINNIMFGILNISLLDGFVSILASATSQQC---KRNSSFSLEGTNFTFSDTRNK 118
Query: 379 FTALGCNVVAMLLNGSTGYSGGCASFCSSGNNIIDGSCSGVACCQAPVPKGLKKLDLEFS 558
FTALGC++VAMLLNGS+GYSGGCASFCS+ +NIIDG CSGVACCQAPVPKGLKKL+LEF+
Sbjct: 119 FTALGCDMVAMLLNGSSGYSGGCASFCSTKSNIIDGMCSGVACCQAPVPKGLKKLELEFT 178
Query: 559 NIN--ITNSGIDYDYWPCGQAFVVEQDSYVFSALDLNNTNDTKPQYRHVVLEWSVNGSSC 732
NI ++ + CG+AF+VEQ+SYVFS++DL+NTN PQYR VVLEWS++G C
Sbjct: 179 NITGQLSRPKEVNNTPTCGEAFIVEQNSYVFSSVDLSNTNRNNPQYRPVVLEWSIDGGYC 238
Query: 733 EEAKLSGSYACAENAYCYNSSNGIGYRCNCSSGFEGNPYLQGPNGC 870
EEA SYAC EN+YCYNSSNGIGYRCNCS GF+GNPYLQGP+GC
Sbjct: 239 EEANRFMSYACKENSYCYNSSNGIGYRCNCSLGFQGNPYLQGPDGC 284
>sptr|Q7XNT7|Q7XNT7 OSJNBa0093F12.21 protein.
Length = 707
Score = 206 bits (523), Expect = 7e-52
Identities = 113/283 (39%), Positives = 155/283 (54%), Gaps = 17/283 (6%)
Frame = +1
Query: 55 LTCFSPSFVHPMEPGTYRCSNVSIPIPYPFGVAGKSPSLSR-GFKVVCGHSGPLLSIGNS 231
L+ S S + P+ +C N S+ IP+PF +A S S GF + C +GP++ +G +
Sbjct: 18 LSSSSSSQISPL-----KCPNSSVDIPFPFKIATNSSLTSTPGFAISCRQTGPMILLGGN 72
Query: 232 MXXXXXXXXXXXXXXXXASASSQQCIGGTANSSVNLEGTVFTFSDTRNKFTALGCNVVAM 411
+ S QC + ++L T + FS T+NKFTA+GC+ +AM
Sbjct: 73 YSVLSISLLEGYVRVTGQTVYSSQC-HNNSQGIIDLTATNYMFSHTQNKFTAVGCDAMAM 131
Query: 412 LLNGS-----------TGYSGGCASFCSSGNNIIDGSCSGVACCQAPVPKGLKKLDLEFS 558
+ N S + YSGGC SFC+S +II G CSGV CCQ+ VPKGL KLDLEF+
Sbjct: 132 IRNSSDVVGNTNSTVMSRYSGGCVSFCASNGSIISGECSGVGCCQSSVPKGLNKLDLEFT 191
Query: 559 NIN----ITNSGIDYDYWPCGQAFVVEQDSYVFSALDLNNTNDTKPQYRHVVLEWSVNGS 726
+I S + C +AF+ EQDSYVFS DL P +VL+W + G
Sbjct: 192 SIRDQLMPPTSAVGSGSTRCSKAFIAEQDSYVFSRHDLYKDLGNLP----MVLDWYIQGG 247
Query: 727 SCEEAKLS-GSYACAENAYCYNSSNGIGYRCNCSSGFEGNPYL 852
+C+EA S +Y C EN+YCY +G GYRCNCS G+ GNPY+
Sbjct: 248 NCKEASRSRQTYMCKENSYCYEVEDGAGYRCNCSGGYTGNPYI 290
>sptr|Q7XKW0|Q7XKW0 OSJNBa0073E02.9 protein.
Length = 742
Score = 181 bits (459), Expect = 2e-44
Identities = 111/298 (37%), Positives = 156/298 (52%), Gaps = 13/298 (4%)
Frame = +1
Query: 106 RCSNVSIPIPYPFGVAGKSPSLSRGFKVVC----GHSGPLLSIGNSMXXXXXXXXXXXXX 273
RC +V I YPFG+ G + SL+ F V C G S P + +
Sbjct: 33 RCGDVDIQ--YPFGI-GANCSLAELFNVECKVQHGISKPFIGNVEVLNISLSRSTLRVLN 89
Query: 274 XXXASASSQQCIGGTANSSVNLEGTVFTFSDTRNKFTALGCNVVAMLLN-GSTGYSGGCA 450
+ + G + N + T F FSD NKFT +GCN +A + + G TGY GC
Sbjct: 90 GISTFCYNASGLMGGVHFRFNAKNTPFRFSDVYNKFTVIGCNTLAYIADDGGTGYQSGCF 149
Query: 451 SFCSSGNNIIDGSCSGVACCQAPVPKGLKKLDLEFSNINITNSGIDYDYWPCGQAFVVEQ 630
S C + ++DGSCSG+ CCQ +P+G+ ++ F S I + C A ++E
Sbjct: 150 SQCRDLSGLVDGSCSGMGCCQTTIPRGMYYYNVTFDK-RFNTSQISR-FGRCSYAVLMEA 207
Query: 631 DSYVFSALDLNNT--NDTKPQYRHVVLEWSVNGSSCEEAKLS-GSYAC-AENAYCYNSSN 798
S+ FS +N T N T +V++W++ SC+ AK + SYAC + N+ C S+N
Sbjct: 208 ASFNFSTTYINTTKFNGTNGGRVPMVIDWAIREKSCDIAKQNMTSYACLSSNSECVASTN 267
Query: 799 GIGYRCNCSSGFEGNPYLQGPNGCTDINECITRN--PCTNR--CSNTIGGFQCTCPAG 960
G GY CNCS G+EGNPYL P+GC D+NEC RN PC + C NT GG++C+C G
Sbjct: 268 GPGYVCNCSHGYEGNPYLPDPHGCHDVNEC-DRNPWPCPSGGVCHNTEGGYRCSCRKG 324
>sptrnew|CAD40634|CAD40634 OSJNBa0016N04.10 protein.
Length = 739
Score = 180 bits (457), Expect = 3e-44
Identities = 96/225 (42%), Positives = 133/225 (59%), Gaps = 11/225 (4%)
Frame = +1
Query: 331 VNLEGTVFTFSDTRNKFTALGCNVVAMLLNGST--GYSGGCASFCSSGN--NIIDGSCSG 498
+N+ GT F SD+ NKFT +GC A + + Y GC S C G+ +G+CSG
Sbjct: 113 LNMTGTPFMLSDS-NKFTVVGCRTQAYIADQDYVGKYMSGCVSVCRRGDVWKATNGTCSG 171
Query: 499 VACCQAPVPKGLKKLDLEFSNINITNSGIDYDYWPCGQAFVVEQDSYVFSALDLNNT--N 672
+ CCQ +PKGL F + ++ SGI Y+ PC A +++ ++ FS L + N
Sbjct: 172 IGCCQTAIPKGLDYYQAFFDDSSMNTSGI-YNRTPCSYAVLMDSSNFTFSTTYLTTSEFN 230
Query: 673 DTKPQYRHVVLEWSV-NGSSCEEA-KLSGSYAC-AENAYCYNSSNGIGYRCNCSSGFEGN 843
+T +VL+W++ + +SCEEA K SYAC + N+ C+NS+NG GY CNCS G+EGN
Sbjct: 231 NTYDGRAPMVLDWAIRSANSCEEAWKKMDSYACKSTNSECFNSTNGPGYTCNCSKGYEGN 290
Query: 844 PYLQGPNGCTDINEC--ITRNPCTNRCSNTIGGFQCTCPAGMSGD 972
PYL+GPNGC DI+EC + C C N GGF C CPAG G+
Sbjct: 291 PYLEGPNGCRDIDECQDSKTHHCYGECRNKPGGFDCNCPAGSKGN 335
>sptr|Q7XVH8|Q7XVH8 OSJNBa0016N04.12 protein.
Length = 739
Score = 180 bits (457), Expect = 3e-44
Identities = 96/225 (42%), Positives = 133/225 (59%), Gaps = 11/225 (4%)
Frame = +1
Query: 331 VNLEGTVFTFSDTRNKFTALGCNVVAMLLNGST--GYSGGCASFCSSGN--NIIDGSCSG 498
+N+ GT F SD+ NKFT +GC A + + Y GC S C G+ +G+CSG
Sbjct: 113 LNMTGTPFMLSDS-NKFTVVGCRTQAYIADQDYVGKYMSGCVSVCRRGDVWKATNGTCSG 171
Query: 499 VACCQAPVPKGLKKLDLEFSNINITNSGIDYDYWPCGQAFVVEQDSYVFSALDLNNT--N 672
+ CCQ +PKGL F + ++ SGI Y+ PC A +++ ++ FS L + N
Sbjct: 172 IGCCQTAIPKGLDYYQAFFDDSSMNTSGI-YNRTPCSYAVLMDSSNFTFSTTYLTTSEFN 230
Query: 673 DTKPQYRHVVLEWSV-NGSSCEEA-KLSGSYAC-AENAYCYNSSNGIGYRCNCSSGFEGN 843
+T +VL+W++ + +SCEEA K SYAC + N+ C+NS+NG GY CNCS G+EGN
Sbjct: 231 NTYDGRAPMVLDWAIRSANSCEEAWKKMDSYACKSTNSECFNSTNGPGYTCNCSKGYEGN 290
Query: 844 PYLQGPNGCTDINEC--ITRNPCTNRCSNTIGGFQCTCPAGMSGD 972
PYL+GPNGC DI+EC + C C N GGF C CPAG G+
Sbjct: 291 PYLEGPNGCRDIDECQDSKTHHCYGECRNKPGGFDCNCPAGSKGN 335
>sptrnew|CAE05044|CAE05044 OSJNBa0049H08.5 protein.
Length = 676
Score = 177 bits (450), Expect = 2e-43
Identities = 95/230 (41%), Positives = 134/230 (58%), Gaps = 14/230 (6%)
Frame = +1
Query: 325 SSVNLEGTVFTFSDTRNKFTALGCNVVAMLLNGSTGYSG-----GCASFCSSGNNIID-- 483
++++L G+ FTFS++ NKFT GC ++ L G G GCA+ C G++++
Sbjct: 21 ANLSLTGSPFTFSNSANKFTVFGCRMLGYLGPGGQSAVGSNLTIGCATSCGQGDDLLSIN 80
Query: 484 -GSCSGVACCQAPVPKGLKKLDLEFSNINITNSGIDYDYWPCGQAFVVEQDSYVFSALDL 660
CSG+ CCQ +PKG+K + F N+ + +++ C +VE+ S+ FS +
Sbjct: 81 GEGCSGIGCCQTAIPKGIKHYKVWFDTH--FNTSVIHNWSRCSYGALVEEASFKFSTIYA 138
Query: 661 NNTNDTKPQYRH--VVLEWSVNGSSCEEA-KLSGSYACAE-NAYCYNSSNGIGYRCNCSS 828
++N + P V++W V ++C EA K SYACA N+ C +SSNG GY C CS
Sbjct: 139 TSSNFSNPFGGEPPFVVDWVVANNTCAEARKHLDSYACASSNSVCIDSSNGPGYFCKCSQ 198
Query: 829 GFEGNPYLQGPNGCTDINECITRN--PCTNRCSNTIGGFQCTCPAGMSGD 972
GFEGNPYLQG +GC DINEC N PC +C N +GGF C CPAG GD
Sbjct: 199 GFEGNPYLQGHDGCQDINECEDSNKYPCYGKCINKLGGFDCFCPAGTRGD 248
>sptrnew|CAD40762|CAD40762 OSJNBa0081G05.15 protein.
Length = 676
Score = 177 bits (450), Expect = 2e-43
Identities = 95/230 (41%), Positives = 134/230 (58%), Gaps = 14/230 (6%)
Frame = +1
Query: 325 SSVNLEGTVFTFSDTRNKFTALGCNVVAMLLNGSTGYSG-----GCASFCSSGNNIID-- 483
++++L G+ FTFS++ NKFT GC ++ L G G GCA+ C G++++
Sbjct: 21 ANLSLTGSPFTFSNSANKFTVFGCRMLGYLGPGGQSAVGSNLTIGCATSCGQGDDLLSIN 80
Query: 484 -GSCSGVACCQAPVPKGLKKLDLEFSNINITNSGIDYDYWPCGQAFVVEQDSYVFSALDL 660
CSG+ CCQ +PKG+K + F N+ + +++ C +VE+ S+ FS +
Sbjct: 81 GEGCSGIGCCQTAIPKGIKHYKVWFDTH--FNTSVIHNWSRCSYGALVEEASFKFSTIYA 138
Query: 661 NNTNDTKPQYRH--VVLEWSVNGSSCEEA-KLSGSYACAE-NAYCYNSSNGIGYRCNCSS 828
++N + P V++W V ++C EA K SYACA N+ C +SSNG GY C CS
Sbjct: 139 TSSNFSNPFGGEPPFVVDWVVANNTCAEARKHLDSYACASSNSVCIDSSNGPGYFCKCSQ 198
Query: 829 GFEGNPYLQGPNGCTDINECITRN--PCTNRCSNTIGGFQCTCPAGMSGD 972
GFEGNPYLQG +GC DINEC N PC +C N +GGF C CPAG GD
Sbjct: 199 GFEGNPYLQGHDGCQDINECEDSNKYPCYGKCINKLGGFDCFCPAGTRGD 248
>sptr|Q7X8L4|Q7X8L4 OSJNBa0081G05.22 protein (OSJNBa0049H08.6
protein).
Length = 667
Score = 177 bits (450), Expect = 2e-43
Identities = 95/230 (41%), Positives = 134/230 (58%), Gaps = 14/230 (6%)
Frame = +1
Query: 325 SSVNLEGTVFTFSDTRNKFTALGCNVVAMLLNGSTGYSG-----GCASFCSSGNNIID-- 483
++++L G+ FTFS++ NKFT GC ++ L G G GCA+ C G++++
Sbjct: 12 ANLSLTGSPFTFSNSANKFTVFGCRMLGYLGPGGQSAVGSNLTIGCATSCGQGDDLLSIN 71
Query: 484 -GSCSGVACCQAPVPKGLKKLDLEFSNINITNSGIDYDYWPCGQAFVVEQDSYVFSALDL 660
CSG+ CCQ +PKG+K + F N+ + +++ C +VE+ S+ FS +
Sbjct: 72 GEGCSGIGCCQTAIPKGIKHYKVWFDTH--FNTSVIHNWSRCSYGALVEEASFKFSTIYA 129
Query: 661 NNTNDTKPQYRH--VVLEWSVNGSSCEEA-KLSGSYACAE-NAYCYNSSNGIGYRCNCSS 828
++N + P V++W V ++C EA K SYACA N+ C +SSNG GY C CS
Sbjct: 130 TSSNFSNPFGGEPPFVVDWVVANNTCAEARKHLDSYACASSNSVCIDSSNGPGYFCKCSQ 189
Query: 829 GFEGNPYLQGPNGCTDINECITRN--PCTNRCSNTIGGFQCTCPAGMSGD 972
GFEGNPYLQG +GC DINEC N PC +C N +GGF C CPAG GD
Sbjct: 190 GFEGNPYLQGHDGCQDINECEDSNKYPCYGKCINKLGGFDCFCPAGTRGD 239
>sptr|Q7XVA3|Q7XVA3 OSJNBa0081G05.12 protein.
Length = 591
Score = 177 bits (449), Expect = 3e-43
Identities = 112/312 (35%), Positives = 161/312 (51%), Gaps = 29/312 (9%)
Frame = +1
Query: 124 IPIPYPFGVAGKSPSLSR-GFKVVCGH---SGPLLSIGN----SMXXXXXXXXXXXXXXX 279
I +P+PFG+ G P ++ GF++ CG+ SG + + S
Sbjct: 10 IEVPFPFGIDGDQPGCAKPGFELSCGNNTESGVPILLRKVQPLSRSVEVLGISLPKGQLR 69
Query: 280 XASASSQQCIGGTA-------NSSVNLEGTVFTFSDTRNKFTALGCNVVAMLLNGSTGYS 438
S C T N ++L G+ FTFSD+ NKFTA GC V+A L G
Sbjct: 70 MRMHMSSHCYNMTTRVMDCVDNGWMDLTGSPFTFSDSANKFTAFGCQVLAYLGAGEQRDI 129
Query: 439 G-----GCASFCSSGNN--IIDGSCSGVACCQAPVPKGLKKLDLEFSNINITNSGIDYDY 597
G GCA+ C ++ I G CSG+ CCQ +PKG+K F + T+S Y +
Sbjct: 130 GSNLRIGCAASCGKDDSATIGGGRCSGIGCCQTAIPKGIKYYKAWFDDRFNTSSM--YTW 187
Query: 598 WPCGQAFVVEQDSYVFSALDLNNT---NDTKPQYRHVVLEWSVNGSSCEEAKLS-GSYAC 765
C A +VE+ S+ FS + +++ +DT V++W + SC+EA+ + G+Y C
Sbjct: 188 NRCAYAALVEESSFNFSMIYDSSSKFNSDTVSSQPPFVVDWVMGNISCKEARKNLGTYPC 247
Query: 766 -AENAYCYNSSNGIGYRCNCSSGFEGNPYLQGPNGCTDINEC--ITRNPCTNRCSNTIGG 936
+ N+ C +S NG GY CNC GF+GNPY +G + C DINEC + PC +C N +GG
Sbjct: 248 ISNNSICLDSQNGPGYICNCRKGFQGNPYNKGLDSCQDINECDDPKKYPCYGKCINKLGG 307
Query: 937 FQCTCPAGMSGD 972
F C CPAGM G+
Sbjct: 308 FDCFCPAGMRGN 319
>sptr|Q7XH96|Q7XH96 Putative wall-associated protein kinase.
Length = 793
Score = 170 bits (431), Expect = 3e-41
Identities = 113/326 (34%), Positives = 152/326 (46%), Gaps = 33/326 (10%)
Frame = +1
Query: 106 RCSNVSIPIPYPFGVAGKSPSLSRGFKVVCGHSG----PLLSIGNSMXXXXXXXXXXXXX 273
RC +V IP YPFG+ G S GF++ C S P L+ N
Sbjct: 32 RCGDVDIP--YPFGIGGGC-FRSAGFEIACNTSNGGLVPTLAAANDTIQVQNLTVFPRPE 88
Query: 274 XXXASASSQQCIGGTANSSVNLEGTV-------FTFSDTRNKFTALGCNVVAMLLNGSTG 432
+ +C + N + G V + SD RNKF LGCN VA +G +
Sbjct: 89 VKVMLPVAYRCYNSSGNVTEQFYGDVELNKTGVYRISDERNKFVVLGCNTVAWNKHGDSE 148
Query: 433 --------YSGGCASFCSSGNNIIDGSCSGVACCQAPVPKGLKKLDLEFSNINITNSGID 588
Y GC ++CS + +G C+GV CC +P L + F G
Sbjct: 149 GKGLYTSLYYAGCVTYCSDSLSAKNGKCAGVGCCHVDIPPELTDNVVTFQQ---WPRGEQ 205
Query: 589 YDYWPCGQAFVVEQDSYVFSALDLNNTNDTKPQYRHVVLEWSVNG-SSCEEAKLSGS--- 756
D+ PC AF+V+++ Y F DLN + Q V L+W++ +SC ++ S
Sbjct: 206 VDFSPCDYAFLVDKEEYQFQRSDLNMD---RKQRMPVWLDWAIRDVASCPAPEVETSKKN 262
Query: 757 ----YACAE-NAYCYNSSNGIGYRCNCSSGFEGNPYLQGPN-GCTDINECITRN--PCTN 912
YAC N+ C NS+NG+GY CNCSSG+EGNPY PN GC DI+EC N PC
Sbjct: 263 MPAGYACVSVNSTCVNSTNGLGYYCNCSSGYEGNPYDDDPNKGCKDIDECAHPNKYPCHG 322
Query: 913 RCSNTIGGFQCTCPAGM--SGDGLKE 984
C NT G ++C C G SGDG K+
Sbjct: 323 VCRNTPGDYECRCHTGYQPSGDGPKK 348
>sptr|Q94LI8|Q94LI8 Putative wall-associated protein kinase.
Length = 793
Score = 170 bits (431), Expect = 3e-41
Identities = 113/326 (34%), Positives = 152/326 (46%), Gaps = 33/326 (10%)
Frame = +1
Query: 106 RCSNVSIPIPYPFGVAGKSPSLSRGFKVVCGHSG----PLLSIGNSMXXXXXXXXXXXXX 273
RC +V IP YPFG+ G S GF++ C S P L+ N
Sbjct: 32 RCGDVDIP--YPFGIGGGC-FRSAGFEIACNTSNGGLVPTLAAANDTIQVQNLTVFPRPE 88
Query: 274 XXXASASSQQCIGGTANSSVNLEGTV-------FTFSDTRNKFTALGCNVVAMLLNGSTG 432
+ +C + N + G V + SD RNKF LGCN VA +G +
Sbjct: 89 VKVMLPVAYRCYNSSGNVTEQFYGDVELNKTGVYRISDERNKFVVLGCNTVAWNKHGDSE 148
Query: 433 --------YSGGCASFCSSGNNIIDGSCSGVACCQAPVPKGLKKLDLEFSNINITNSGID 588
Y GC ++CS + +G C+GV CC +P L + F G
Sbjct: 149 GKGLYTSLYYAGCVTYCSDSLSAKNGKCAGVGCCHVDIPPELTDNVVTFQQ---WPRGEQ 205
Query: 589 YDYWPCGQAFVVEQDSYVFSALDLNNTNDTKPQYRHVVLEWSVNG-SSCEEAKLSGS--- 756
D+ PC AF+V+++ Y F DLN + Q V L+W++ +SC ++ S
Sbjct: 206 VDFSPCDYAFLVDKEEYQFQRSDLNMD---RKQRMPVWLDWAIRDVASCPAPEVETSKKN 262
Query: 757 ----YACAE-NAYCYNSSNGIGYRCNCSSGFEGNPYLQGPN-GCTDINECITRN--PCTN 912
YAC N+ C NS+NG+GY CNCSSG+EGNPY PN GC DI+EC N PC
Sbjct: 263 MPAGYACVSVNSTCVNSTNGLGYYCNCSSGYEGNPYDDDPNKGCKDIDECAHPNKYPCHG 322
Query: 913 RCSNTIGGFQCTCPAGM--SGDGLKE 984
C NT G ++C C G SGDG K+
Sbjct: 323 VCRNTPGDYECRCHTGYQPSGDGPKK 348
>sptr|Q7XGR6|Q7XGR6 Putative wall-associated protein kinase.
Length = 748
Score = 166 bits (419), Expect = 8e-40
Identities = 109/328 (33%), Positives = 150/328 (45%), Gaps = 38/328 (11%)
Frame = +1
Query: 106 RCSNVSIPIPYPFGVAGKSPSLSRGFKVVC--------GHSGPLLSIGNSMXXXXXXXXX 261
RC +VSIP YPFG+ G + S +GF++ C G P L+ N
Sbjct: 31 RCGDVSIP--YPFGI-GPNCSHGKGFEIACDTRTRNGSGELVPTLAAANGTIHVQSLFVA 87
Query: 262 XXXXXXXASASSQQCIGGTANSSVNLEGTV-------FTFSDTRNKFTALGCNVVAMLLN 420
+ QC + + + + G V + SD RN F LGCN +A N
Sbjct: 88 PIPEVKVMLPVAYQCYNSSDSVTESFFGAVDLNNNGVYRISDKRNMFVVLGCNTMAYTNN 147
Query: 421 GSTGYSG--------GCASFCSSGNNIIDGSCSGVACCQAPVPKGLKKLDLEFSNINITN 576
G + G GC S+C+ ++ DG C+G+ CC + GL + F +
Sbjct: 148 GDSHGKGPYAGLYYTGCVSYCNDSSSAQDGMCAGIGCCHVDISPGLSDNVVTFGE---WS 204
Query: 577 SGIDYDYWPCGQAFVVEQDSYVFSALDLN-NTNDTKPQYRHVVLEWSVN-------GSSC 732
D+ PC AF+V +D Y F DL + N TKP V L+W++ SSC
Sbjct: 205 RYFQVDFNPCNYAFLVAKDEYNFQRSDLQKDLNRTKP----VWLDWAIRDGGNSSASSSC 260
Query: 733 E----EAKLSGSYAC-AENAYCYNSSNGIGYRCNCSSGFEGNPYLQGPNGCTDINECITR 897
K+ YAC ++N+ C NS+NG GY C CS G+EGNPYL G GC DI+EC
Sbjct: 261 PAPEVREKMPPEYACVSDNSECVNSTNGPGYYCKCSKGYEGNPYLVG--GCNDIDECARS 318
Query: 898 N--PCTNRCSNTIGGFQCTCPAGMSGDG 975
+ PC C NT+G + C C G G
Sbjct: 319 DEYPCHGDCRNTVGDYHCKCRTGYQPRG 346
>sptr|Q8W2R7|Q8W2R7 Putative wall-associated protein kinase.
Length = 748
Score = 166 bits (419), Expect = 8e-40
Identities = 109/328 (33%), Positives = 150/328 (45%), Gaps = 38/328 (11%)
Frame = +1
Query: 106 RCSNVSIPIPYPFGVAGKSPSLSRGFKVVC--------GHSGPLLSIGNSMXXXXXXXXX 261
RC +VSIP YPFG+ G + S +GF++ C G P L+ N
Sbjct: 31 RCGDVSIP--YPFGI-GPNCSHGKGFEIACDTRTRNGSGELVPTLAAANGTIHVQSLFVA 87
Query: 262 XXXXXXXASASSQQCIGGTANSSVNLEGTV-------FTFSDTRNKFTALGCNVVAMLLN 420
+ QC + + + + G V + SD RN F LGCN +A N
Sbjct: 88 PIPEVKVMLPVAYQCYNSSDSVTESFFGAVDLNNNGVYRISDKRNMFVVLGCNTMAYTNN 147
Query: 421 GSTGYSG--------GCASFCSSGNNIIDGSCSGVACCQAPVPKGLKKLDLEFSNINITN 576
G + G GC S+C+ ++ DG C+G+ CC + GL + F +
Sbjct: 148 GDSHGKGPYAGLYYTGCVSYCNDSSSAQDGMCAGIGCCHVDISPGLSDNVVTFGE---WS 204
Query: 577 SGIDYDYWPCGQAFVVEQDSYVFSALDLN-NTNDTKPQYRHVVLEWSVN-------GSSC 732
D+ PC AF+V +D Y F DL + N TKP V L+W++ SSC
Sbjct: 205 RYFQVDFNPCNYAFLVAKDEYNFQRSDLQKDLNRTKP----VWLDWAIRDGGNSSASSSC 260
Query: 733 E----EAKLSGSYAC-AENAYCYNSSNGIGYRCNCSSGFEGNPYLQGPNGCTDINECITR 897
K+ YAC ++N+ C NS+NG GY C CS G+EGNPYL G GC DI+EC
Sbjct: 261 PAPEVREKMPPEYACVSDNSECVNSTNGPGYYCKCSKGYEGNPYLVG--GCNDIDECARS 318
Query: 898 N--PCTNRCSNTIGGFQCTCPAGMSGDG 975
+ PC C NT+G + C C G G
Sbjct: 319 DEYPCHGDCRNTVGDYHCKCRTGYQPRG 346
>sptr|Q7XGS0|Q7XGS0 Putative wall-associated protein kinase.
Length = 693
Score = 166 bits (419), Expect = 8e-40
Identities = 107/328 (32%), Positives = 157/328 (47%), Gaps = 35/328 (10%)
Frame = +1
Query: 106 RCSNVSIPIPYPFGVAGKSPSLSRGFKVVC------GHSGPLLSIGNSMXXXXXXXXXXX 267
RC ++ IP YPFG+ G + S +GF++ C G P L+ N
Sbjct: 31 RCGDIDIP--YPFGI-GPNCSRGKGFEIACNPRNDSGEMVPTLAAANGTIHVQSLLVAPI 87
Query: 268 XXXXXASASSQQC------IGGTANSSVNLEGT-VFTFSDTRNKFTALGCNVVAMLLNGS 426
+ QC I + V+L T V+ SD+RN F +GCN ++ NG+
Sbjct: 88 PEVKVMLPVAYQCYYSNNSITDSFYGEVDLNNTGVYRISDSRNMFVVIGCNTLSYTQNGN 147
Query: 427 TGYSG--------GCASFCSSGNNIIDGSCSGVACCQAPVPKGLKKLDLEFSNINITNSG 582
+G G GC S+C+ ++ D C+GV CC + GL + F G
Sbjct: 148 SGGKGPYAGLYYTGCVSYCNDSSSARDSMCAGVGCCHIDISPGLSDNVVSFGP---WKRG 204
Query: 583 IDYDYWPCGQAFVVEQDSYVFSALDLN-NTNDTKPQYRHVVLEWSVNGS------SCEEA 741
D+ PC +F+V+++ Y F + DL + N T P V L+W++ S +E
Sbjct: 205 FQVDFSPCDYSFLVDKNEYEFRSADLKMDLNRTMP----VWLDWAIRDSVTCPPLEVQEK 260
Query: 742 KLSGSYAC-AENAYCYNSSNGIGYRCNCSSGFEGNPYLQGPNGCTDINECITRN----PC 906
K +G YAC ++N+ C NS+NG GY C C G++GNPY+ GC DINEC N PC
Sbjct: 261 KPAG-YACMSDNSECVNSTNGPGYYCKCKQGYDGNPYVDKDQGCKDINECDVSNKKKYPC 319
Query: 907 TNRCSNTIGGFQCTCPAG--MSGDGLKE 984
C+N G ++C C G SG+G K+
Sbjct: 320 YGVCNNIPGDYECHCRVGYQWSGEGPKK 347
>sptr|Q94H08|Q94H08 Putative wall-associated protein kinase.
Length = 693
Score = 166 bits (419), Expect = 8e-40
Identities = 107/328 (32%), Positives = 157/328 (47%), Gaps = 35/328 (10%)
Frame = +1
Query: 106 RCSNVSIPIPYPFGVAGKSPSLSRGFKVVC------GHSGPLLSIGNSMXXXXXXXXXXX 267
RC ++ IP YPFG+ G + S +GF++ C G P L+ N
Sbjct: 31 RCGDIDIP--YPFGI-GPNCSRGKGFEIACNPRNDSGEMVPTLAAANGTIHVQSLLVAPI 87
Query: 268 XXXXXASASSQQC------IGGTANSSVNLEGT-VFTFSDTRNKFTALGCNVVAMLLNGS 426
+ QC I + V+L T V+ SD+RN F +GCN ++ NG+
Sbjct: 88 PEVKVMLPVAYQCYYSNNSITDSFYGEVDLNNTGVYRISDSRNMFVVIGCNTLSYTQNGN 147
Query: 427 TGYSG--------GCASFCSSGNNIIDGSCSGVACCQAPVPKGLKKLDLEFSNINITNSG 582
+G G GC S+C+ ++ D C+GV CC + GL + F G
Sbjct: 148 SGGKGPYAGLYYTGCVSYCNDSSSARDSMCAGVGCCHIDISPGLSDNVVSFGP---WKRG 204
Query: 583 IDYDYWPCGQAFVVEQDSYVFSALDLN-NTNDTKPQYRHVVLEWSVNGS------SCEEA 741
D+ PC +F+V+++ Y F + DL + N T P V L+W++ S +E
Sbjct: 205 FQVDFSPCDYSFLVDKNEYEFRSADLKMDLNRTMP----VWLDWAIRDSVTCPPLEVQEK 260
Query: 742 KLSGSYAC-AENAYCYNSSNGIGYRCNCSSGFEGNPYLQGPNGCTDINECITRN----PC 906
K +G YAC ++N+ C NS+NG GY C C G++GNPY+ GC DINEC N PC
Sbjct: 261 KPAG-YACMSDNSECVNSTNGPGYYCKCKQGYDGNPYVDKDQGCKDINECDVSNKKKYPC 319
Query: 907 TNRCSNTIGGFQCTCPAG--MSGDGLKE 984
C+N G ++C C G SG+G K+
Sbjct: 320 YGVCNNIPGDYECHCRVGYQWSGEGPKK 347
>sptrnew|CAD40704|CAD40704 OSJNBb0042I07.1 protein.
Length = 766
Score = 164 bits (414), Expect = 3e-39
Identities = 103/315 (32%), Positives = 155/315 (49%), Gaps = 25/315 (7%)
Frame = +1
Query: 85 PMEPGTYRCSNVSIPIPYPFGVAGKSPS---LSRGFKVVCGHSGPLLSIGNSMXXXXXXX 255
P PG + + IPYPFGV S S GF + C +
Sbjct: 38 PPPPGNCQRKCGDVDIPYPFGVWNGSESDGCAVPGFYLNCDVDDNHVYRPFHGNVEVLSI 97
Query: 256 XXXXXXXXXASASSQQCIGGTANS------SVNLEGTVFTFSDTRNKFTALGCNVVAMLL 417
++ S C ++ +N GT T SD NKFT +GC +A +
Sbjct: 98 SLPTGQARVTNSISSACYNTSSRDMDYNDWQINFTGTPLTISDADNKFTVVGCQTLAYIT 157
Query: 418 NGST--GYSGGCASFCSSGNNI---IDGSCSGVACCQAPVPKGLKKLDLEFSNINITNSG 582
+ Y+ GC + C G+ +GSCSG+ CCQ +P+GLK + F N+
Sbjct: 158 DDDNMGKYTSGCVAMCQGGDLTSLATNGSCSGIGCCQTAIPRGLKYYRVRFDTG--FNTS 215
Query: 583 IDYDYWPCGQAFVVEQDSYVF-----SALDLNNTNDTKPQYRHVVLEWSVNGSSCEEAKL 747
Y+ C A ++E ++ F S+L+ N++N + +V++W++ +C++A+
Sbjct: 216 EIYNVSRCSYAVLMESKAFSFRTSYVSSLEFNSSNGGRVP---LVVDWAIGNETCDKARR 272
Query: 748 S-GSYAC-AENAYCYNSSNGIGYRCNCSSGFEGNPYLQ-GPNGCTDINECI-TRNPCT-- 909
+YAC + N+ C+NSSNG GY CNCS G++GNPYLQ G +GCTDI+EC + PC+
Sbjct: 273 KVDTYACVSHNSKCFNSSNGPGYICNCSEGYQGNPYLQDGQHGCTDIDECADPKYPCSVP 332
Query: 910 NRCSNTIGGFQCTCP 954
C N GGF+C CP
Sbjct: 333 GTCHNLPGGFECLCP 347
>sptrnew|CAD40631|CAD40631 OSJNBa0016N04.13 protein.
Length = 766
Score = 164 bits (414), Expect = 3e-39
Identities = 103/315 (32%), Positives = 155/315 (49%), Gaps = 25/315 (7%)
Frame = +1
Query: 85 PMEPGTYRCSNVSIPIPYPFGVAGKSPS---LSRGFKVVCGHSGPLLSIGNSMXXXXXXX 255
P PG + + IPYPFGV S S GF + C +
Sbjct: 38 PPPPGNCQRKCGDVDIPYPFGVWNGSESDGCAVPGFYLNCDVDDNHVYRPFHGNVEVLSI 97
Query: 256 XXXXXXXXXASASSQQCIGGTANS------SVNLEGTVFTFSDTRNKFTALGCNVVAMLL 417
++ S C ++ +N GT T SD NKFT +GC +A +
Sbjct: 98 SLPTGQARVTNSISSACYNTSSRDMDYNDWQINFTGTPLTISDADNKFTVVGCQTLAYIT 157
Query: 418 NGST--GYSGGCASFCSSGNNI---IDGSCSGVACCQAPVPKGLKKLDLEFSNINITNSG 582
+ Y+ GC + C G+ +GSCSG+ CCQ +P+GLK + F N+
Sbjct: 158 DDDNMGKYTSGCVAMCQGGDLTSLATNGSCSGIGCCQTAIPRGLKYYRVRFDTG--FNTS 215
Query: 583 IDYDYWPCGQAFVVEQDSYVF-----SALDLNNTNDTKPQYRHVVLEWSVNGSSCEEAKL 747
Y+ C A ++E ++ F S+L+ N++N + +V++W++ +C++A+
Sbjct: 216 EIYNVSRCSYAVLMESKAFSFRTSYVSSLEFNSSNGGRVP---LVVDWAIGNETCDKARR 272
Query: 748 S-GSYAC-AENAYCYNSSNGIGYRCNCSSGFEGNPYLQ-GPNGCTDINECI-TRNPCT-- 909
+YAC + N+ C+NSSNG GY CNCS G++GNPYLQ G +GCTDI+EC + PC+
Sbjct: 273 KVDTYACVSHNSKCFNSSNGPGYICNCSEGYQGNPYLQDGQHGCTDIDECADPKYPCSVP 332
Query: 910 NRCSNTIGGFQCTCP 954
C N GGF+C CP
Sbjct: 333 GTCHNLPGGFECLCP 347
>sptr|Q7X895|Q7X895 OSJNBa0016N04.17 protein (OSJNBb0042I07.2
protein).
Length = 766
Score = 164 bits (414), Expect = 3e-39
Identities = 103/315 (32%), Positives = 155/315 (49%), Gaps = 25/315 (7%)
Frame = +1
Query: 85 PMEPGTYRCSNVSIPIPYPFGVAGKSPS---LSRGFKVVCGHSGPLLSIGNSMXXXXXXX 255
P PG + + IPYPFGV S S GF + C +
Sbjct: 38 PPPPGNCQRKCGDVDIPYPFGVWNGSESDGCAVPGFYLNCDVDDNHVYRPFHGNVEVLSI 97
Query: 256 XXXXXXXXXASASSQQCIGGTANS------SVNLEGTVFTFSDTRNKFTALGCNVVAMLL 417
++ S C ++ +N GT T SD NKFT +GC +A +
Sbjct: 98 SLPTGQARVTNSISSACYNTSSRDMDYNDWQINFTGTPLTISDADNKFTVVGCQTLAYIT 157
Query: 418 NGST--GYSGGCASFCSSGNNI---IDGSCSGVACCQAPVPKGLKKLDLEFSNINITNSG 582
+ Y+ GC + C G+ +GSCSG+ CCQ +P+GLK + F N+
Sbjct: 158 DDDNMGKYTSGCVAMCQGGDLTSLATNGSCSGIGCCQTAIPRGLKYYRVRFDTG--FNTS 215
Query: 583 IDYDYWPCGQAFVVEQDSYVF-----SALDLNNTNDTKPQYRHVVLEWSVNGSSCEEAKL 747
Y+ C A ++E ++ F S+L+ N++N + +V++W++ +C++A+
Sbjct: 216 EIYNVSRCSYAVLMESKAFSFRTSYVSSLEFNSSNGGRVP---LVVDWAIGNETCDKARR 272
Query: 748 S-GSYAC-AENAYCYNSSNGIGYRCNCSSGFEGNPYLQ-GPNGCTDINECI-TRNPCT-- 909
+YAC + N+ C+NSSNG GY CNCS G++GNPYLQ G +GCTDI+EC + PC+
Sbjct: 273 KVDTYACVSHNSKCFNSSNGPGYICNCSEGYQGNPYLQDGQHGCTDIDECADPKYPCSVP 332
Query: 910 NRCSNTIGGFQCTCP 954
C N GGF+C CP
Sbjct: 333 GTCHNLPGGFECLCP 347
>sptr|Q7XGR2|Q7XGR2 Putative wall-associated protein kinase.
Length = 695
Score = 160 bits (405), Expect = 3e-38
Identities = 108/329 (32%), Positives = 154/329 (46%), Gaps = 36/329 (10%)
Frame = +1
Query: 106 RCSNVSIPIPYPFGVAGKSPSLSRGFKVVC------GHSGPLLSIGNSMXXXXXXXXXXX 267
RC +V IP YPFG+ G + S GF++ C G P L+ N
Sbjct: 33 RCGDVDIP--YPFGI-GPNCSRGEGFEIACNTRNGNGDLVPTLAAANGSIHVQSLSVEQL 89
Query: 268 XXXXXASASSQQCIGGTANSSVNLEGTV-------FTFSDTRNKFTALGCNVVAMLLNGS 426
+ +C N + G V + SD+RN F +GCN ++ NG+
Sbjct: 90 PEVKVMLPVAYKCYDAGDNVTRRFYGDVDLNNNGVYRISDSRNMFVVIGCNTLSYTQNGN 149
Query: 427 TG-----YSG----GCASFCSSGNNIIDGSCSGVACCQAPVPKGLKKLDLEFSNINITNS 579
+G YSG GC ++C+ + DG C+GV CC + GL + F
Sbjct: 150 SGGSNTHYSGLFYTGCVTYCNDSRSAQDGRCAGVGCCHVDISPGLTDNVVSFGP---WTR 206
Query: 580 GIDYDYWPCGQAFVVEQDSYVFSALDLN-NTNDTKPQYRHVVLEWSVNGS------SCEE 738
G D+ PC +F+V+++ Y F + DL + N T P V L+W++ S +E
Sbjct: 207 GFQVDFSPCDYSFLVDKNEYEFRSADLKMDLNRTMP----VWLDWAIRDSVTCPPLEVQE 262
Query: 739 AKLSGSYAC-AENAYCYNSSNGIGYRCNCSSGFEGNPYLQGPNGCTDINECITRN----P 903
K +G YAC ++N+ C NS+NG GY C C G+EGNPY GC DINEC N P
Sbjct: 263 KKPAG-YACVSDNSECVNSTNGPGYYCKCKQGYEGNPY-DKDQGCKDINECDVSNKKKYP 320
Query: 904 CTNRCSNTIGGFQCTCPAG--MSGDGLKE 984
C C+N G ++C C G SG+G K+
Sbjct: 321 CYGVCNNIPGDYECHCRVGYQWSGEGPKK 349
>sptr|Q8W2R3|Q8W2R3 Putative wall-associated protein kinase.
Length = 695
Score = 160 bits (405), Expect = 3e-38
Identities = 108/329 (32%), Positives = 154/329 (46%), Gaps = 36/329 (10%)
Frame = +1
Query: 106 RCSNVSIPIPYPFGVAGKSPSLSRGFKVVC------GHSGPLLSIGNSMXXXXXXXXXXX 267
RC +V IP YPFG+ G + S GF++ C G P L+ N
Sbjct: 33 RCGDVDIP--YPFGI-GPNCSRGEGFEIACNTRNGNGDLVPTLAAANGSIHVQSLSVEQL 89
Query: 268 XXXXXASASSQQCIGGTANSSVNLEGTV-------FTFSDTRNKFTALGCNVVAMLLNGS 426
+ +C N + G V + SD+RN F +GCN ++ NG+
Sbjct: 90 PEVKVMLPVAYKCYDAGDNVTRRFYGDVDLNNNGVYRISDSRNMFVVIGCNTLSYTQNGN 149
Query: 427 TG-----YSG----GCASFCSSGNNIIDGSCSGVACCQAPVPKGLKKLDLEFSNINITNS 579
+G YSG GC ++C+ + DG C+GV CC + GL + F
Sbjct: 150 SGGSNTHYSGLFYTGCVTYCNDSRSAQDGRCAGVGCCHVDISPGLTDNVVSFGP---WTR 206
Query: 580 GIDYDYWPCGQAFVVEQDSYVFSALDLN-NTNDTKPQYRHVVLEWSVNGS------SCEE 738
G D+ PC +F+V+++ Y F + DL + N T P V L+W++ S +E
Sbjct: 207 GFQVDFSPCDYSFLVDKNEYEFRSADLKMDLNRTMP----VWLDWAIRDSVTCPPLEVQE 262
Query: 739 AKLSGSYAC-AENAYCYNSSNGIGYRCNCSSGFEGNPYLQGPNGCTDINECITRN----P 903
K +G YAC ++N+ C NS+NG GY C C G+EGNPY GC DINEC N P
Sbjct: 263 KKPAG-YACVSDNSECVNSTNGPGYYCKCKQGYEGNPY-DKDQGCKDINECDVSNKKKYP 320
Query: 904 CTNRCSNTIGGFQCTCPAG--MSGDGLKE 984
C C+N G ++C C G SG+G K+
Sbjct: 321 CYGVCNNIPGDYECHCRVGYQWSGEGPKK 349
>sptrnew|AAQ56778|AAQ56778 At1g21270.
Length = 732
Score = 160 bits (404), Expect = 4e-38
Identities = 94/216 (43%), Positives = 125/216 (57%), Gaps = 7/216 (3%)
Frame = +1
Query: 352 FTFSDTRNKFTALGCNVVAML-LNGSTGYSGGCASFCSSGNNIIDGSCSGVACCQAPVPK 528
FT S+ N+FT +GCN A L +G YS GC S C S +GSCSG CCQ PVP+
Sbjct: 112 FTLSEL-NRFTVVGCNSYAFLRTSGVEKYSTGCISICDSATTK-NGSCSGEGCCQIPVPR 169
Query: 529 GLKKLDLEFSNINITNSGIDYDYWPCGQAFVVEQDSYVFSAL-DLNNTND--TKPQYRHV 699
G + ++ + + N + + PC AF+VE + F AL DLNN + T P V
Sbjct: 170 GYSFVRVKPHSFH--NHPTVHLFNPCTYAFLVEDGMFDFHALEDLNNLRNVTTFP----V 223
Query: 700 VLEWSVNGSSCEEAKLSGSYACAENAYCYNSSNGIGYRCNCSSGFEGNPYLQGPNGCTDI 879
VL+WS+ +C++ + G C N+ C++S+ G GY C C GFEGNPYL PNGC DI
Sbjct: 224 VLDWSIGDKTCKQVEYRG--VCGGNSTCFDSTGGTGYNCKCLEGFEGNPYL--PNGCQDI 279
Query: 880 NECI-TRNPCT--NRCSNTIGGFQCTCPAGMSGDGL 978
NECI +R+ C+ + C NT G F C CP+G D L
Sbjct: 280 NECISSRHNCSEHSTCENTKGSFNCNCPSGYRKDSL 315
>sptr|Q9XGN2|Q9XGN2 Wall-associated kinase 2 (Hypothetical protein).
Length = 732
Score = 160 bits (404), Expect = 4e-38
Identities = 94/216 (43%), Positives = 125/216 (57%), Gaps = 7/216 (3%)
Frame = +1
Query: 352 FTFSDTRNKFTALGCNVVAML-LNGSTGYSGGCASFCSSGNNIIDGSCSGVACCQAPVPK 528
FT S+ N+FT +GCN A L +G YS GC S C S +GSCSG CCQ PVP+
Sbjct: 112 FTLSEL-NRFTVVGCNSYAFLRTSGVEKYSTGCISICDSATTK-NGSCSGEGCCQIPVPR 169
Query: 529 GLKKLDLEFSNINITNSGIDYDYWPCGQAFVVEQDSYVFSAL-DLNNTND--TKPQYRHV 699
G + ++ + + N + + PC AF+VE + F AL DLNN + T P V
Sbjct: 170 GYSFVRVKPHSFH--NHPTVHLFNPCTYAFLVEDGMFDFHALEDLNNLRNVTTFP----V 223
Query: 700 VLEWSVNGSSCEEAKLSGSYACAENAYCYNSSNGIGYRCNCSSGFEGNPYLQGPNGCTDI 879
VL+WS+ +C++ + G C N+ C++S+ G GY C C GFEGNPYL PNGC DI
Sbjct: 224 VLDWSIGDKTCKQVEYRG--VCGGNSTCFDSTGGTGYNCKCLEGFEGNPYL--PNGCQDI 279
Query: 880 NECI-TRNPCT--NRCSNTIGGFQCTCPAGMSGDGL 978
NECI +R+ C+ + C NT G F C CP+G D L
Sbjct: 280 NECISSRHNCSEHSTCENTKGSFNCNCPSGYRKDSL 315
>sptr|Q9LMP1|Q9LMP1 F16F4.5 protein (Wall-associated kinase 2).
Length = 732
Score = 160 bits (404), Expect = 4e-38
Identities = 94/216 (43%), Positives = 125/216 (57%), Gaps = 7/216 (3%)
Frame = +1
Query: 352 FTFSDTRNKFTALGCNVVAML-LNGSTGYSGGCASFCSSGNNIIDGSCSGVACCQAPVPK 528
FT S+ N+FT +GCN A L +G YS GC S C S +GSCSG CCQ PVP+
Sbjct: 112 FTLSEL-NRFTVVGCNSYAFLRTSGVEKYSTGCISICDSATTK-NGSCSGEGCCQIPVPR 169
Query: 529 GLKKLDLEFSNINITNSGIDYDYWPCGQAFVVEQDSYVFSAL-DLNNTND--TKPQYRHV 699
G + ++ + + N + + PC AF+VE + F AL DLNN + T P V
Sbjct: 170 GYSFVRVKPHSFH--NHPTVHLFNPCTYAFLVEDGMFDFHALEDLNNLRNVTTFP----V 223
Query: 700 VLEWSVNGSSCEEAKLSGSYACAENAYCYNSSNGIGYRCNCSSGFEGNPYLQGPNGCTDI 879
VL+WS+ +C++ + G C N+ C++S+ G GY C C GFEGNPYL PNGC DI
Sbjct: 224 VLDWSIGDKTCKQVEYRG--VCGGNSTCFDSTGGTGYNCKCLEGFEGNPYL--PNGCQDI 279
Query: 880 NECI-TRNPCT--NRCSNTIGGFQCTCPAGMSGDGL 978
NECI +R+ C+ + C NT G F C CP+G D L
Sbjct: 280 NECISSRHNCSEHSTCENTKGSFNCNCPSGYRKDSL 315
>sptrnew|CAD40632|CAD40632 OSJNBa0016N04.12 protein.
Length = 732
Score = 157 bits (398), Expect = 2e-37
Identities = 103/309 (33%), Positives = 150/309 (48%), Gaps = 18/309 (5%)
Frame = +1
Query: 124 IPIPYPFGVAGKSPSLSRGFKVVCGHSGP------LLSIGNSMXXXXXXXXXXXXXXXXA 285
I IPYPFG+ + + C + P +LSI +
Sbjct: 38 IDIPYPFGIGSDGDCALPFYNIDCNNKKPFYRDVEVLSISLQLGQIRVSTPISSSCYNPF 97
Query: 286 SASSQQCIGGTANSSVNLEGTVFTFSDTRNKFTALGCNVVAMLLNGSTGYSGGCASFCSS 465
S G NL T F SD+ NKFT +GC +A + + ++ Y+ GCAS C
Sbjct: 98 SKRMYSSGWG-----FNLSYTPFMLSDS-NKFTVVGCQSLAYISDPTSNYTSGCASSCPG 151
Query: 466 GNNI--IDGSCSGVACCQAPVPKGLKKLDLEFSNINITNSGIDYDYWPCGQAFVVEQDSY 639
G + + +CS + CCQ +P+G++ + F ++ SGI Y++ PC A +++ ++
Sbjct: 152 GKVVSATNRTCSRIGCCQITIPRGMEFCKVSFGE-SMNTSGI-YEHTPCSYAAIMDYSNF 209
Query: 640 VFSALDLNNT---NDTKPQYRHVVLEWSVNGS-SCEEAKLS-GSYAC-AENAYCYNSSNG 801
FS +L + N+T V +W++ G C EA+ + SYAC ++++ C N S+G
Sbjct: 210 TFSTSNLTSLLEFNNTYSGRAPVKFDWAIWGPRDCVEAQKNLTSYACKSDHSVCLNYSSG 269
Query: 802 I--GYRCNCSSGFEGNPYLQGPNGCTDINEC--ITRNPCTNRCSNTIGGFQCTCPAGMSG 969
Y CNCS G+ GNPYLQG NGC DINEC PC C N GGF C C G G
Sbjct: 270 AKSAYMCNCSKGYHGNPYLQGSNGCEDINECEHPESYPCYGECHNKDGGFDCFCRDGTRG 329
Query: 970 DGLKEGSGC 996
+ G GC
Sbjct: 330 NATIPG-GC 337
>sptr|Q7XVI1|Q7XVI1 OSJNBa0016N04.15 protein.
Length = 707
Score = 154 bits (390), Expect = 2e-36
Identities = 99/298 (33%), Positives = 146/298 (48%), Gaps = 18/298 (6%)
Frame = +1
Query: 124 IPIPYPFGVAGKSPSLSRGFKVVCGHSGP------LLSIGNSMXXXXXXXXXXXXXXXXA 285
I IPYPFG+ + + C + P +LSI +
Sbjct: 38 IDIPYPFGIGSDGDCALPFYNIDCNNKKPFYRDVEVLSISLQLGQIRVSTPISSSCYNPF 97
Query: 286 SASSQQCIGGTANSSVNLEGTVFTFSDTRNKFTALGCNVVAMLLNGSTGYSGGCASFCSS 465
S G NL T F SD+ NKFT +GC +A + + ++ Y+ GCAS C
Sbjct: 98 SKRMYSSGWG-----FNLSYTPFMLSDS-NKFTVVGCQSLAYISDPTSNYTSGCASSCPG 151
Query: 466 GNNI--IDGSCSGVACCQAPVPKGLKKLDLEFSNINITNSGIDYDYWPCGQAFVVEQDSY 639
G + + +CS + CCQ +P+G++ + F ++ SGI Y++ PC A +++ ++
Sbjct: 152 GKVVSATNRTCSRIGCCQITIPRGMEFCKVSFGE-SMNTSGI-YEHTPCSYAAIMDYSNF 209
Query: 640 VFSALDLNNT---NDTKPQYRHVVLEWSVNGS-SCEEAKLS-GSYAC-AENAYCYNSSNG 801
FS +L + N+T V +W++ G C EA+ + SYAC ++++ C N S+G
Sbjct: 210 TFSTSNLTSLLEFNNTYSGRAPVKFDWAIWGPRDCVEAQKNLTSYACKSDHSVCLNYSSG 269
Query: 802 I--GYRCNCSSGFEGNPYLQGPNGCTDINEC--ITRNPCTNRCSNTIGGFQCTCPAGM 963
Y CNCS G+ GNPYLQG NGC DINEC PC C N GGF C C G+
Sbjct: 270 AKSAYMCNCSKGYHGNPYLQGSNGCEDINECEHPESYPCYGECHNKDGGFDCFCRDGV 327
>sptr|Q7XLN2|Q7XLN2 OSJNBa0049H08.9 protein.
Length = 657
Score = 154 bits (390), Expect = 2e-36
Identities = 95/269 (35%), Positives = 137/269 (50%), Gaps = 20/269 (7%)
Frame = +1
Query: 124 IPIPYPFGVAGKSPSLSRGFKVVCGHSG---PLLSIGN----SMXXXXXXXXXXXXXXXX 282
I IP+PFG+AG+ GFK+ C +G P L + N +
Sbjct: 19 IDIPFPFGIAGQPGCAMTGFKLSCNDTGNGVPTLLLRNVEVLGISLPLGQARMKMDMSYD 78
Query: 283 ASASSQQCIGGTANSSVNLEGTVFTFSDTRNKFTALGCNVVAMLLNGSTGYSG-----GC 447
++++ I +NL+G+ FTFSDT NKF GC ++A L G G GC
Sbjct: 79 CYNTTRKDIDCVDMVDLNLKGSPFTFSDTANKFIVFGCRMLAYLGPGEQNDVGSNLTIGC 138
Query: 448 ASFCSSGNNIID---GSCSGVACCQAPVPKGLKKLDLEFSN-INITNSGIDYDYWPCGQA 615
A+ C G++++ CSG+ CCQ +PKG++ + F N T+ Y++ C A
Sbjct: 139 AATCGIGDDLVSINSAGCSGIGCCQTNIPKGIRYYKVWFDGRFNTTDI---YNWTRCAYA 195
Query: 616 FVVEQDSYVFSAL--DLNNTNDTKPQYRHVVLEWSVNGSSCEEAKL-SGSYAC-AENAYC 783
+VE S+ FS + L+ ND V++W++ S+CE+AK S SY C + N+ C
Sbjct: 196 ALVETSSFNFSTVYNSLSRFNDNLGSQPPFVVDWAIGNSTCEQAKTNSDSYMCISSNSVC 255
Query: 784 YNSSNGIGYRCNCSSGFEGNPYLQGPNGC 870
NS NG GY CNC +GFEGNPYL GC
Sbjct: 256 LNSRNGPGYICNCQNGFEGNPYLNDSFGC 284
>sptr|Q9SNQ7|Q9SNQ7 Similar to wak1 gene.
Length = 734
Score = 154 bits (389), Expect = 2e-36
Identities = 105/313 (33%), Positives = 148/313 (47%), Gaps = 27/313 (8%)
Frame = +1
Query: 94 PGTYRCSNVSIPIPYPFGVAGKSPSLSRGFKVVC----GHSGPLLSIGNSMXXXXXXXXX 261
P +C NV+IP YPFG+ GF + C + GP + + N
Sbjct: 40 PAPAKCGNVNIP--YPFGIREGCFRPKGGFNISCKQEQAYIGPDIRVTNFDVVQSEARIL 97
Query: 262 XXXXXXXAS---ASSQQCIGGTANSSVNLEGTVFTFSDTRNKFTALGCNVVAMLL----N 420
+ + I T+ + L G S +N+FTA+GC+ VA + N
Sbjct: 98 TDIPSGTVAWKYNNEFDPIAWTSRGGLRL-GNHHMVSSAKNRFTAIGCSTVAFIYGRDKN 156
Query: 421 GSTG----YSGGCASFCSSGNNIIDG-SCSGVACCQAPVPKGLKKLDLEFSNINITNSGI 585
GS G ++ C SFC +I DG CSG+ CCQ P+ L++ L F N N T +
Sbjct: 157 GSNGQFDQFTSLCGSFCFDEGSIEDGPECSGMGCCQVPISTNLRRFSLGFYNYNTTKKVL 216
Query: 586 DYDYWPCGQAFVVEQDSYVFSALDLNNTNDTKPQYRHV--VLEWSVNGSSCEEAKLSGSY 759
++ AFVVE+D + F + N + R + +L+W +C+EA L SY
Sbjct: 217 NFS--SRSYAFVVEKDQFKFKSSYAKADNFMEELARGIPIILQWIAGNETCKEAALKESY 274
Query: 760 AC-AENAYCYNSSNGIGYRCNCSSGFEGNPYLQGPNGC-----TDINEC-ITR--NPCTN 912
AC A N+ C + GYRCNC+ G+EGNPYL+ +GC DINEC TR N C
Sbjct: 275 ACVANNSKCIDVIEAPGYRCNCTQGYEGNPYLK--DGCRVVNIADINECNATRFPNSCKG 332
Query: 913 RCSNTIGGFQCTC 951
C+NT G + C C
Sbjct: 333 ICTNTDGSYDCIC 345
>sptr|Q9ARW4|Q9ARW4 Putative wall-associated kinase 2.
Length = 725
Score = 152 bits (385), Expect = 7e-36
Identities = 96/302 (31%), Positives = 148/302 (49%), Gaps = 12/302 (3%)
Frame = +1
Query: 130 IPYPFGVAGKSPSLSRGFKVVCGHSGPLLSIGNSMXXXXXXXXXXXXXXXXASASSQQCI 309
IPYPFG+ G ++ GF + C + + +A S C
Sbjct: 35 IPYPFGI-GTECAIEPGFVIYCNKTADGSMKPFLINVEVLNISLLHGQTRALNALSTYCY 93
Query: 310 GGTANS------SVNLEGTVFTFSDTRNKFTALGCNVVAMLLNGSTGYSGGCASFCSSGN 471
S S++ + FS+ NKF +GCN ++ + NG Y+ CAS C+
Sbjct: 94 NDVTKSMESSRWSLDFSTWPYRFSNLHNKFVVIGCNTLSYIYNGE--YTTACASVCAKAP 151
Query: 472 NIIDGSCSGVACCQAPVPKGLKKLDLEFSNINITNSGIDYDYWPCGQAFVVEQDSYVFSA 651
+ SC GV CCQ + KGL ++ F + +S + + PC A +VE D++ F
Sbjct: 152 T--NDSCDGVGCCQNNIAKGLNSYNVTFFTVYNDSSNLQSN--PCSYAALVETDTFRFKT 207
Query: 652 --LDLNNTNDTKPQYRHVVLEWSVNGSSCEEAKLSGSYAC-AENAYCYNSSNGIGYRCNC 822
+ N+T + VVL+W++ C+EA ++ SYAC ++++ C +S NG GY CNC
Sbjct: 208 EYVTTMKFNETYNGQQPVVLDWAIGKVGCKEANMT-SYACRSKHSECVDSINGPGYLCNC 266
Query: 823 SSGFEGNPYLQGPNGCTDINEC-ITRNPCTN--RCSNTIGGFQCTCPAGMSGDGLKEGSG 993
+ G+ GNPY+ +GC D+NEC ++PC C NT G + C+CP G KE +
Sbjct: 267 TLGYHGNPYI--TDGCIDVNECEQNQSPCPKGATCRNTEGWYHCSCPVGRK--LAKETNT 322
Query: 994 CN 999
CN
Sbjct: 323 CN 324
>sptrnew|CAD40714|CAD40714 OSJNBb0042I07.11 protein.
Length = 771
Score = 149 bits (375), Expect = 1e-34
Identities = 98/301 (32%), Positives = 152/301 (50%), Gaps = 22/301 (7%)
Frame = +1
Query: 34 LWHIPFFL-TCFSPSFVHPMEPGTYRCSNVS----IPIPYPFGVAGKSPSLSRGFKVVCG 198
+W + L T + + + + +C N + + I YPFG++ ++S F+V C
Sbjct: 20 IWLLVLLLSTALAAETLDGAQAASSQCQNATKCGGVDIVYPFGLSSSGCAMSPSFEVDCN 79
Query: 199 HSGPLLS---IGNSMXXXXXXXXXXXXXXXXASASSQQCIGGTANSS----VNLEGTVFT 357
++G + +G S+S N V+L+ T +
Sbjct: 80 NTGNGVQKPFLGYVELLSIDVQLSQARVRTRISSSCYNISTREMNFDDLWYVDLKDTPYR 139
Query: 358 FSDTRNKFTALGCNVVAMLLNGST--GYSGGCASFCSSGN--NIIDGSCSGVACCQAPVP 525
FSD+ NKFT +GC +A + + Y GC S C G ++I+G+CSG CCQ +P
Sbjct: 140 FSDSANKFTIIGCRTLAYIADQDDVGKYMSGCVSVCRRGELTSLINGTCSGKGCCQTAIP 199
Query: 526 KGLKKLDLEFSNINITNSGIDYDYWPCGQAFVVEQDSYVFSALDLNNT---NDTKPQYRH 696
KGL + F ++ SGI Y+ PC A ++E ++ FS L + N+T
Sbjct: 200 KGLDYYQVWFEQ-SMNTSGI-YNRTPCSYAVLMEASNFSFSTTYLTSPFEFNNTYGGEAP 257
Query: 697 VVLEWSVN-GSSCEEAKLS-GSYAC-AENAYCYNSSNGIGYRCNCSSGFEGNPYLQGPNG 867
VVL+W++N ++CEEA + SYAC ++NA C NSS+ GY C C G++GNPYL+GPNG
Sbjct: 258 VVLDWAINTANTCEEAMGNLTSYACKSDNAKCINSSDTTGYICRCQEGYQGNPYLKGPNG 317
Query: 868 C 870
C
Sbjct: 318 C 318
Database: swall
Posted date: Feb 26, 2004 3:00 PM
Number of letters in database: 439,479,560
Number of sequences in database: 1,381,838
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 987,300,489
Number of Sequences: 1381838
Number of extensions: 23327707
Number of successful extensions: 74494
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 61514
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 71294
length of database: 439,479,560
effective HSP length: 125
effective length of database: 266,749,810
effective search space used: 66420702690
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

