BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
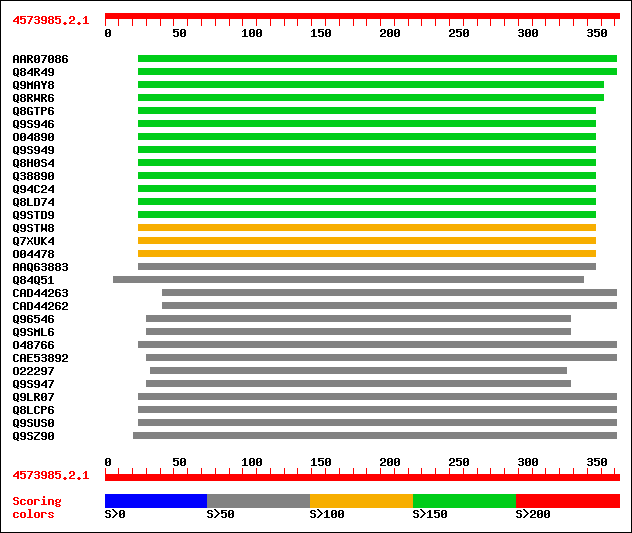
Query= 4573985.2.1
(373 letters)
Database: /db/uniprot/tmp/swall
1,395,590 sequences; 442,889,342 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sptrnew|AAR07086|AAR07086 Putative endo-1,4-beta-glucanase. 198 2e-50
sptr|Q84R49|Q84R49 Putative endo-1,4-beta-glucanase. 198 2e-50
sptr|Q9MAY8|Q9MAY8 Endo-1,4-beta-glucanase Cel1. 187 3e-47
sptr|Q8RWR6|Q8RWR6 Endo-1,4-beta-glucanase. 187 3e-47
sptr|Q8GTP6|Q8GTP6 Endo-1,4-beta-D-glucanase (EC 3.2.1.4). 169 1e-41
sptr|Q9S946|Q9S946 CEL6=CELLULASE 6 (Fragment). 168 1e-41
sptr|O04890|O04890 Endo-1,4-beta-glucanase (EC 3.2.1.4). 167 2e-41
sptr|Q9S949|Q9S949 CEL3=CELLULASE 3 (Fragment). 167 3e-41
sptr|Q8H0S4|Q8H0S4 At5g49720/K2I5_8. 155 2e-37
sptr|Q38890|Q38890 Cellulase precursor (Cellulase homolog OR16PE... 155 2e-37
sptr|Q94C24|Q94C24 AT5g49720/K2I5_8. 155 2e-37
sptr|Q8LD74|Q8LD74 Cellulase homolog OR16pep. 155 2e-37
sptr|Q9STD9|Q9STD9 Cellulase (EC 3.2.1.4). 154 3e-37
sptr|Q9STW8|Q9STW8 Endo-1, 4-beta-glucanase like protein. 140 4e-33
sptr|Q7XUK4|Q7XUK4 OSJNBa0067K08.14 protein. 124 3e-28
sptr|O04478|O04478 F5I14.14. 106 8e-23
sptrnew|AAQ63883|AAQ63883 Cellulase. 86 1e-16
sptr|Q84Q51|Q84Q51 P0456B03.7 protein. 67 4e-11
sptrnew|CAD44263|CAD44263 Putative endo-1,4-beta-glucanase (Frag... 65 2e-10
sptrnew|CAD44262|CAD44262 Putative endo-1,4-beta-glucanase (Frag... 65 2e-10
sptr|Q96546|Q96546 Endo-beta-1,4-glucanase (EC 3.2.1.4). 65 3e-10
sptr|Q9SML6|Q9SML6 Endo-beta-1,4-glucanase precursor (EC 3.2.1.4). 64 6e-10
sptr|O48766|O48766 Putative glucanse (At2g32990). 63 8e-10
sptrnew|CAE53892|CAE53892 Putative endo-1,3(4)-beta-glucanase (F... 61 3e-09
sptr|O22297|O22297 Acidic cellulase. 61 4e-09
sptr|Q9S947|Q9S947 CEL5=CELLULASE 5 (Fragment). 60 7e-09
sptr|Q9LR07|Q9LR07 F10A5.13 (Hypothetical protein) (Putative end... 60 9e-09
sptr|Q8LCP6|Q8LCP6 Endo-beta-1,4-glucanase, putative. 60 9e-09
sptr|Q9SUS0|Q9SUS0 Putative cellulase. 60 9e-09
sptr|Q9SZ90|Q9SZ90 Cellulase-like protein. 60 9e-09
>sptrnew|AAR07086|AAR07086 Putative endo-1,4-beta-glucanase.
Length = 620
Score = 198 bits (503), Expect = 2e-50
Identities = 94/120 (78%), Positives = 101/120 (84%), Gaps = 4/120 (3%)
Frame = -3
Query: 371 AWS----RPRYEAIGELDHIKELIKWGTDYILKTFNSSADTIDRIVAQVGVGDTSKGSSQ 204
+WS + +YEAIGELDH+KELIKWGTDY+LKTFNSSADTIDRIVAQVGVGDTSKG +Q
Sbjct: 181 SWSVIEYKAKYEAIGELDHVKELIKWGTDYLLKTFNSSADTIDRIVAQVGVGDTSKGGAQ 240
Query: 203 PNDHYCWMRPEDIDYKRPVTECHSCSDXXXXXXXXXXXXSIVFKDSKTYSDKLVKGAKAL 24
PNDHYCWMRPEDIDY RPVTECHSCSD SIVFKDSKTYSDKLV+GAKAL
Sbjct: 241 PNDHYCWMRPEDIDYPRPVTECHSCSDLASEMAAALAAASIVFKDSKTYSDKLVRGAKAL 300
>sptr|Q84R49|Q84R49 Putative endo-1,4-beta-glucanase.
Length = 620
Score = 198 bits (503), Expect = 2e-50
Identities = 94/120 (78%), Positives = 101/120 (84%), Gaps = 4/120 (3%)
Frame = -3
Query: 371 AWS----RPRYEAIGELDHIKELIKWGTDYILKTFNSSADTIDRIVAQVGVGDTSKGSSQ 204
+WS + +YEAIGELDH+KELIKWGTDY+LKTFNSSADTIDRIVAQVGVGDTSKG +Q
Sbjct: 181 SWSVIEYKAKYEAIGELDHVKELIKWGTDYLLKTFNSSADTIDRIVAQVGVGDTSKGGAQ 240
Query: 203 PNDHYCWMRPEDIDYKRPVTECHSCSDXXXXXXXXXXXXSIVFKDSKTYSDKLVKGAKAL 24
PNDHYCWMRPEDIDY RPVTECHSCSD SIVFKDSKTYSDKLV+GAKAL
Sbjct: 241 PNDHYCWMRPEDIDYPRPVTECHSCSDLASEMAAALAAASIVFKDSKTYSDKLVRGAKAL 300
>sptr|Q9MAY8|Q9MAY8 Endo-1,4-beta-glucanase Cel1.
Length = 621
Score = 187 bits (475), Expect = 3e-47
Identities = 90/113 (79%), Positives = 94/113 (83%)
Frame = -3
Query: 362 RPRYEAIGELDHIKELIKWGTDYILKTFNSSADTIDRIVAQVGVGDTSKGSSQPNDHYCW 183
R +YEAIGELDH+KE+IKWGTDY+LKTFNSSADTIDRIVAQVGVGDTSKG PNDHYCW
Sbjct: 189 RAKYEAIGELDHVKEIIKWGTDYMLKTFNSSADTIDRIVAQVGVGDTSKGP-MPNDHYCW 247
Query: 182 MRPEDIDYKRPVTECHSCSDXXXXXXXXXXXXSIVFKDSKTYSDKLVKGAKAL 24
MRPEDIDYKRPV ECHSCSD SIVFKDSK YSDKLV GAKAL
Sbjct: 248 MRPEDIDYKRPVIECHSCSDLAAEMAAALAAASIVFKDSKAYSDKLVHGAKAL 300
>sptr|Q8RWR6|Q8RWR6 Endo-1,4-beta-glucanase.
Length = 620
Score = 187 bits (475), Expect = 3e-47
Identities = 90/113 (79%), Positives = 94/113 (83%)
Frame = -3
Query: 362 RPRYEAIGELDHIKELIKWGTDYILKTFNSSADTIDRIVAQVGVGDTSKGSSQPNDHYCW 183
R +YEAIGELDH+KE+IKWGTDY+LKTFNSSADTIDRIVAQVGVGDTSKG PNDHYCW
Sbjct: 189 RAKYEAIGELDHVKEIIKWGTDYMLKTFNSSADTIDRIVAQVGVGDTSKGP-MPNDHYCW 247
Query: 182 MRPEDIDYKRPVTECHSCSDXXXXXXXXXXXXSIVFKDSKTYSDKLVKGAKAL 24
MRPEDIDYKRPV ECHSCSD SIVFKDSK YSDKLV GAKAL
Sbjct: 248 MRPEDIDYKRPVIECHSCSDLAAGMAAALAAASIVFKDSKAYSDKLVHGAKAL 300
>sptr|Q8GTP6|Q8GTP6 Endo-1,4-beta-D-glucanase (EC 3.2.1.4).
Length = 621
Score = 169 bits (427), Expect = 1e-41
Identities = 76/111 (68%), Positives = 88/111 (79%)
Frame = -3
Query: 356 RYEAIGELDHIKELIKWGTDYILKTFNSSADTIDRIVAQVGVGDTSKGSSQPNDHYCWMR 177
+YEA GEL H+KE+IKWG+DY LKTFN SAD+IDR+VAQVGVG T+ GS+ PNDHYCWMR
Sbjct: 192 KYEAAGELSHVKEIIKWGSDYFLKTFNHSADSIDRLVAQVGVGSTAGGSTTPNDHYCWMR 251
Query: 176 PEDIDYKRPVTECHSCSDXXXXXXXXXXXXSIVFKDSKTYSDKLVKGAKAL 24
PEDIDY+RPV++CHSCSD SIVFKD+K YS KLV GAK L
Sbjct: 252 PEDIDYQRPVSQCHSCSDLAAEMAAALAASSIVFKDNKAYSQKLVHGAKTL 302
>sptr|Q9S946|Q9S946 CEL6=CELLULASE 6 (Fragment).
Length = 163
Score = 168 bits (426), Expect = 1e-41
Identities = 78/111 (70%), Positives = 88/111 (79%)
Frame = -3
Query: 356 RYEAIGELDHIKELIKWGTDYILKTFNSSADTIDRIVAQVGVGDTSKGSSQPNDHYCWMR 177
+YEA GEL HIK++IKWGTDY+LKTFNSSADTIDR+V+QVG+GDTS G PNDHYCW+R
Sbjct: 32 KYEAAGELTHIKDVIKWGTDYLLKTFNSSADTIDRLVSQVGIGDTS-GGPDPNDHYCWVR 90
Query: 176 PEDIDYKRPVTECHSCSDXXXXXXXXXXXXSIVFKDSKTYSDKLVKGAKAL 24
PEDIDY RPVTECHSCSD SIVFKD+K YS KLV GA+ L
Sbjct: 91 PEDIDYPRPVTECHSCSDLAAEMAAALASASIVFKDNKVYSQKLVHGARTL 141
>sptr|O04890|O04890 Endo-1,4-beta-glucanase (EC 3.2.1.4).
Length = 617
Score = 167 bits (424), Expect = 2e-41
Identities = 76/111 (68%), Positives = 86/111 (77%)
Frame = -3
Query: 356 RYEAIGELDHIKELIKWGTDYILKTFNSSADTIDRIVAQVGVGDTSKGSSQPNDHYCWMR 177
+YEA GEL H+K+ IKWGTDY+LKTFNSSADTIDRI AQVG GDT+ G++ PNDHYCW+R
Sbjct: 190 KYEAAGELAHVKDTIKWGTDYLLKTFNSSADTIDRIAAQVGKGDTTGGATDPNDHYCWVR 249
Query: 176 PEDIDYKRPVTECHSCSDXXXXXXXXXXXXSIVFKDSKTYSDKLVKGAKAL 24
PEDIDY RPVTECH CSD SIVFKD+K YS KLV GA+ L
Sbjct: 250 PEDIDYARPVTECHGCSDLAAEMAAALASASIVFKDNKAYSQKLVHGARTL 300
>sptr|Q9S949|Q9S949 CEL3=CELLULASE 3 (Fragment).
Length = 176
Score = 167 bits (423), Expect = 3e-41
Identities = 76/111 (68%), Positives = 86/111 (77%)
Frame = -3
Query: 356 RYEAIGELDHIKELIKWGTDYILKTFNSSADTIDRIVAQVGVGDTSKGSSQPNDHYCWMR 177
+YEA GEL H+K+ IKWGTDY+LKTFNSSADTIDRI AQVG GDT+ G++ PNDHYCW+R
Sbjct: 32 KYEAAGELAHVKDTIKWGTDYLLKTFNSSADTIDRIAAQVGKGDTTGGATDPNDHYCWVR 91
Query: 176 PEDIDYKRPVTECHSCSDXXXXXXXXXXXXSIVFKDSKTYSDKLVKGAKAL 24
PEDIDY RPVTECH CSD SIVFKD+K YS KLV GA+ L
Sbjct: 92 PEDIDYARPVTECHGCSDLAAEMARALASASIVFKDNKAYSQKLVHGARTL 142
>sptr|Q8H0S4|Q8H0S4 At5g49720/K2I5_8.
Length = 621
Score = 155 bits (391), Expect = 2e-37
Identities = 72/112 (64%), Positives = 85/112 (75%), Gaps = 1/112 (0%)
Frame = -3
Query: 356 RYEAIGELDHIKELIKWGTDYILKTFNSSADTIDRIVAQVGVGDTSKGSSQPNDHYCWMR 177
+YEA GEL H+KELIKWGTDY LKTFNS+AD+ID +V+QVG G+T G++ PNDHYCWMR
Sbjct: 190 KYEAAGELTHVKELIKWGTDYFLKTFNSTADSIDDLVSQVGSGNTDDGNTDPNDHYCWMR 249
Query: 176 PEDIDYKRPVTECH-SCSDXXXXXXXXXXXXSIVFKDSKTYSDKLVKGAKAL 24
PED+DYKRPVT C+ CSD SIVFKD+K YS KLV GAK +
Sbjct: 250 PEDMDYKRPVTTCNGGCSDLAAEMAAALASASIVFKDNKEYSKKLVHGAKVV 301
>sptr|Q38890|Q38890 Cellulase precursor (Cellulase homolog OR16PEP
precursor).
Length = 621
Score = 155 bits (391), Expect = 2e-37
Identities = 72/112 (64%), Positives = 85/112 (75%), Gaps = 1/112 (0%)
Frame = -3
Query: 356 RYEAIGELDHIKELIKWGTDYILKTFNSSADTIDRIVAQVGVGDTSKGSSQPNDHYCWMR 177
+YEA GEL H+KELIKWGTDY LKTFNS+AD+ID +V+QVG G+T G++ PNDHYCWMR
Sbjct: 190 KYEAAGELTHVKELIKWGTDYFLKTFNSTADSIDDLVSQVGSGNTDDGNTDPNDHYCWMR 249
Query: 176 PEDIDYKRPVTECH-SCSDXXXXXXXXXXXXSIVFKDSKTYSDKLVKGAKAL 24
PED+DYKRPVT C+ CSD SIVFKD+K YS KLV GAK +
Sbjct: 250 PEDMDYKRPVTTCNGGCSDLAAEMAAALASASIVFKDNKEYSKKLVHGAKVV 301
>sptr|Q94C24|Q94C24 AT5g49720/K2I5_8.
Length = 621
Score = 155 bits (391), Expect = 2e-37
Identities = 72/112 (64%), Positives = 85/112 (75%), Gaps = 1/112 (0%)
Frame = -3
Query: 356 RYEAIGELDHIKELIKWGTDYILKTFNSSADTIDRIVAQVGVGDTSKGSSQPNDHYCWMR 177
+YEA GEL H+KELIKWGTDY LKTFNS+AD+ID +V+QVG G+T G++ PNDHYCWMR
Sbjct: 190 KYEAAGELTHVKELIKWGTDYFLKTFNSTADSIDDLVSQVGSGNTDDGNTDPNDHYCWMR 249
Query: 176 PEDIDYKRPVTECH-SCSDXXXXXXXXXXXXSIVFKDSKTYSDKLVKGAKAL 24
PED+DYKRPVT C+ CSD SIVFKD+K YS KLV GAK +
Sbjct: 250 PEDMDYKRPVTTCNGGCSDLAAEMAAALASASIVFKDNKEYSKKLVHGAKVV 301
>sptr|Q8LD74|Q8LD74 Cellulase homolog OR16pep.
Length = 621
Score = 155 bits (391), Expect = 2e-37
Identities = 72/112 (64%), Positives = 85/112 (75%), Gaps = 1/112 (0%)
Frame = -3
Query: 356 RYEAIGELDHIKELIKWGTDYILKTFNSSADTIDRIVAQVGVGDTSKGSSQPNDHYCWMR 177
+YEA GEL H+KELIKWGTDY LKTFNS+AD+ID +V+QVG G+T G++ PNDHYCWMR
Sbjct: 190 KYEAAGELTHVKELIKWGTDYFLKTFNSTADSIDDLVSQVGSGNTDDGNTDPNDHYCWMR 249
Query: 176 PEDIDYKRPVTECH-SCSDXXXXXXXXXXXXSIVFKDSKTYSDKLVKGAKAL 24
PED+DYKRPVT C+ CSD SIVFKD+K YS KLV GAK +
Sbjct: 250 PEDMDYKRPVTTCNGGCSDLAAEMAAALASASIVFKDNKEYSKKLVHGAKVV 301
>sptr|Q9STD9|Q9STD9 Cellulase (EC 3.2.1.4).
Length = 621
Score = 154 bits (389), Expect = 3e-37
Identities = 72/112 (64%), Positives = 85/112 (75%), Gaps = 1/112 (0%)
Frame = -3
Query: 356 RYEAIGELDHIKELIKWGTDYILKTFNSSADTIDRIVAQVGVGDTSKGSSQPNDHYCWMR 177
+YEA GEL H+KELIKWGTDY LKTFNS+AD+ID +V+QVG G+T GS+ PNDHYCWMR
Sbjct: 190 KYEAAGELVHVKELIKWGTDYFLKTFNSTADSIDDLVSQVGSGNTDDGSTDPNDHYCWMR 249
Query: 176 PEDIDYKRPVTECH-SCSDXXXXXXXXXXXXSIVFKDSKTYSDKLVKGAKAL 24
PED+DYKRPVT C+ CSD SIVFKD++ YS KLV GAK +
Sbjct: 250 PEDMDYKRPVTTCNGGCSDLAAEMAAALASASIVFKDNREYSKKLVHGAKTV 301
>sptr|Q9STW8|Q9STW8 Endo-1, 4-beta-glucanase like protein.
Length = 620
Score = 140 bits (353), Expect = 4e-33
Identities = 68/112 (60%), Positives = 80/112 (71%), Gaps = 1/112 (0%)
Frame = -3
Query: 356 RYEAIGELDHIKELIKWGTDYILKTFNSSADTIDRIVAQVGVGDTSKGSSQPNDHYCWMR 177
+Y+A GEL+H+KELIKWGTDY LKTFNSSAD I +V QVG G + +GS NDHYCWMR
Sbjct: 191 KYQAAGELEHVKELIKWGTDYFLKTFNSSADNIYVMVEQVGSGVSGRGSELHNDHYCWMR 250
Query: 176 PEDIDYKRPVTECH-SCSDXXXXXXXXXXXXSIVFKDSKTYSDKLVKGAKAL 24
PEDI YKR V++C+ SCSD SIVFKD++ YS LV GAK L
Sbjct: 251 PEDIHYKRTVSQCYSSCSDLAAEMAAALASASIVFKDNRLYSKNLVHGAKTL 302
>sptr|Q7XUK4|Q7XUK4 OSJNBa0067K08.14 protein.
Length = 623
Score = 124 bits (311), Expect = 3e-28
Identities = 58/111 (52%), Positives = 75/111 (67%)
Frame = -3
Query: 356 RYEAIGELDHIKELIKWGTDYILKTFNSSADTIDRIVAQVGVGDTSKGSSQPNDHYCWMR 177
+Y+A+GE DH++ELIKWGTDY+L TFNSSA TID++ +QVG+ + +QP+DHYCW R
Sbjct: 191 KYKAVGEYDHVRELIKWGTDYLLLTFNSSASTIDKVYSQVGIAKIN--GTQPDDHYCWNR 248
Query: 176 PEDIDYKRPVTECHSCSDXXXXXXXXXXXXSIVFKDSKTYSDKLVKGAKAL 24
PED+ Y RPV S D SIVF+D+ YS KLV GA A+
Sbjct: 249 PEDMAYPRPVQTAGSAPDLGGEMAAALAAASIVFRDNAAYSKKLVNGAAAV 299
>sptr|O04478|O04478 F5I14.14.
Length = 623
Score = 106 bits (264), Expect = 8e-23
Identities = 51/111 (45%), Positives = 68/111 (61%)
Frame = -3
Query: 356 RYEAIGELDHIKELIKWGTDYILKTFNSSADTIDRIVAQVGVGDTSKGSSQPNDHYCWMR 177
+Y+AI E DH+++++KWGTDY+L TFN+SA +D I QVG G + S P+D YCW +
Sbjct: 195 KYKAIDEYDHMRDVLKWGTDYLLLTFNNSATRLDHIYTQVGGG--LRDSESPDDIYCWQK 252
Query: 176 PEDIDYKRPVTECHSCSDXXXXXXXXXXXXSIVFKDSKTYSDKLVKGAKAL 24
PED+ Y RPV S +D SIVF D Y+ KL KGA+ L
Sbjct: 253 PEDMSYDRPVLSSTSAADLGAEVSAALAAASIVFTDKPDYAKKLKKGAETL 303
>sptrnew|AAQ63883|AAQ63883 Cellulase.
Length = 601
Score = 85.9 bits (211), Expect = 1e-16
Identities = 44/112 (39%), Positives = 66/112 (58%), Gaps = 1/112 (0%)
Frame = -3
Query: 356 RYEAIGELDHIKELIKWGTDYILKTFNSSADTIDRIVAQVGVGDTSKGSSQPNDHYCWMR 177
+Y I ELDH++++I+WGTDY+ + F + + +QVG T + +PND CW R
Sbjct: 185 KYADIDELDHVRDIIRWGTDYLHEVFIPPNGSNLTLYSQVG--STISTNHEPNDISCWQR 242
Query: 176 PEDIDYKRPVTECH-SCSDXXXXXXXXXXXXSIVFKDSKTYSDKLVKGAKAL 24
PED+ Y RPV+ C S +D S+VF++ K YS KLV+ A++L
Sbjct: 243 PEDMSYGRPVSVCDGSATDLAGEIVAALSAASMVFEEDKEYSGKLVQAAESL 294
>sptr|Q84Q51|Q84Q51 P0456B03.7 protein.
Length = 529
Score = 67.4 bits (163), Expect = 4e-11
Identities = 46/118 (38%), Positives = 59/118 (50%), Gaps = 4/118 (3%)
Frame = -3
Query: 347 AIGELDHIKELIKWGTDYILKTFNSSADTIDRIVAQVGVGDTSKGSSQPNDHYCWMRPED 168
A GE H+ E IKWGTDY++K ++ D + A+VG GDT DHYCW RPED
Sbjct: 117 AAGERRHVLEAIKWGTDYLVKAHTAA----DELWAEVGDGDT--------DHYCWQRPED 164
Query: 167 IDYKR---PVTECHSCSDXXXXXXXXXXXXSIVFKDSK-TYSDKLVKGAKALSCPGGR 6
+ R V + SD SIVF+ SK YS L++ A+ L G R
Sbjct: 165 MTTSRQAYKVDRDNPGSDVAGETAAALAAASIVFRRSKPRYSRLLLRHAEQLFDFGDR 222
>sptrnew|CAD44263|CAD44263 Putative endo-1,4-beta-glucanase
(Fragment).
Length = 174
Score = 65.1 bits (157), Expect = 2e-10
Identities = 42/117 (35%), Positives = 56/117 (47%), Gaps = 7/117 (5%)
Frame = -3
Query: 371 AWS----RPRYEAIGELDHIKELIKWGTDYILKTFNSSADTIDRIVAQVGVGDTSKGSSQ 204
+WS R + GEL+H E IKWGTDY +K S + + A+VG GDT
Sbjct: 24 SWSVIEFRDQIAEAGELEHALEAIKWGTDYFVKAHTSP----NVLWAEVGDGDT------ 73
Query: 203 PNDHYCWMRPEDIDYKR---PVTECHSCSDXXXXXXXXXXXXSIVFKDSKTYSDKLV 42
DHYCW RPED+ R + E + SD SIVFK + + +L+
Sbjct: 74 --DHYCWQRPEDMTTSRHAYKIDENNPGSDLAGETAAAMAAASIVFKKTNPHYSRLL 128
>sptrnew|CAD44262|CAD44262 Putative endo-1,4-beta-glucanase
(Fragment).
Length = 174
Score = 65.1 bits (157), Expect = 2e-10
Identities = 42/117 (35%), Positives = 56/117 (47%), Gaps = 7/117 (5%)
Frame = -3
Query: 371 AWS----RPRYEAIGELDHIKELIKWGTDYILKTFNSSADTIDRIVAQVGVGDTSKGSSQ 204
+WS R + GEL+H E IKWGTDY +K S + + A+VG GDT
Sbjct: 24 SWSVIEFRDQIAEAGELEHALEAIKWGTDYFVKAHTSP----NVLWAEVGDGDT------ 73
Query: 203 PNDHYCWMRPEDIDYKR---PVTECHSCSDXXXXXXXXXXXXSIVFKDSKTYSDKLV 42
DHYCW RPED+ R + E + SD SIVFK + + +L+
Sbjct: 74 --DHYCWQRPEDMTTSRHAYKIDENNPGSDLAGETAAAMAAASIVFKKTNPHYSRLL 128
>sptr|Q96546|Q96546 Endo-beta-1,4-glucanase (EC 3.2.1.4).
Length = 497
Score = 64.7 bits (156), Expect = 3e-10
Identities = 42/107 (39%), Positives = 56/107 (52%), Gaps = 4/107 (3%)
Frame = -3
Query: 338 ELDHIKELIKWGTDYILKTFNSSADTIDRIVAQVGVGDTSKGSSQPNDHYCWMRPEDIDY 159
+L H KE I+W TDY+LK +S DT+ V VGD ++ DH CW RPED+D
Sbjct: 114 QLGHAKEAIRWSTDYLLKAATASPDTL-----YVQVGDPNQ------DHRCWERPEDMDT 162
Query: 158 KR---PVTECHSCSDXXXXXXXXXXXXSIVFKDS-KTYSDKLVKGAK 30
R V+ + SD SIVFKDS +YS L++ A+
Sbjct: 163 PRNVYKVSPQNPGSDVAAETAAALAAASIVFKDSDPSYSSTLLRTAQ 209
>sptr|Q9SML6|Q9SML6 Endo-beta-1,4-glucanase precursor (EC 3.2.1.4).
Length = 497
Score = 63.5 bits (153), Expect = 6e-10
Identities = 42/107 (39%), Positives = 55/107 (51%), Gaps = 4/107 (3%)
Frame = -3
Query: 338 ELDHIKELIKWGTDYILKTFNSSADTIDRIVAQVGVGDTSKGSSQPNDHYCWMRPEDIDY 159
+L H KE I+W TDY+LK +S DT+ V VGD ++ DH CW RPED+D
Sbjct: 114 QLGHAKEAIRWSTDYLLKAATASPDTL-----YVQVGDPNQ------DHRCWERPEDMDT 162
Query: 158 KR---PVTECHSCSDXXXXXXXXXXXXSIVFKDS-KTYSDKLVKGAK 30
R V+ + SD SIVFKDS +YS L+ A+
Sbjct: 163 PRNVYKVSPQNPGSDVAAETAAALAAASIVFKDSDPSYSSTLLHTAQ 209
>sptr|O48766|O48766 Putative glucanse (At2g32990).
Length = 525
Score = 63.2 bits (152), Expect = 8e-10
Identities = 45/124 (36%), Positives = 58/124 (46%), Gaps = 8/124 (6%)
Frame = -3
Query: 371 AWSRPRY----EAIGELDHIKELIKWGTDYILKTFNSSADTIDRIVAQVGVGDTSKGSSQ 204
+WS Y + GEL H E IKWGTDY +K S + + A+VG GDT
Sbjct: 108 SWSVIEYGDSLASTGELSHALEAIKWGTDYFIKAHTSP----NVLWAEVGDGDT------ 157
Query: 203 PNDHYCWMRPEDIDYKR---PVTECHSCSDXXXXXXXXXXXXSIVFKDSKT-YSDKLVKG 36
DHYCW RPED+ R + E + SD SIVF+ + YS L+
Sbjct: 158 --DHYCWQRPEDMTTSRRAFKIDENNPGSDIAGETAAAMAAASIVFRSTNPHYSHLLLHH 215
Query: 35 AKAL 24
A+ L
Sbjct: 216 AQQL 219
>sptrnew|CAE53892|CAE53892 Putative endo-1,3(4)-beta-glucanase
(Fragment).
Length = 170
Score = 61.2 bits (147), Expect = 3e-09
Identities = 43/120 (35%), Positives = 56/120 (46%), Gaps = 6/120 (5%)
Frame = -3
Query: 371 AWSRPRYEAI--GELDHIKELIKWGTDYILKTFNSSADTIDRIVAQVGVGDTSKGSSQPN 198
AWS + + GEL H ++ ++WG DY+LK + DT+ V VGD K
Sbjct: 32 AWSVLEFGGMMKGELQHARDAVRWGADYLLKA-TAHPDTV-----YVQVGDAGK------ 79
Query: 197 DHYCWMRPEDIDYKRPVTECHSC---SDXXXXXXXXXXXXSIVF-KDSKTYSDKLVKGAK 30
DH CW RPED+D R V + SD S+VF K YS +LV AK
Sbjct: 80 DHACWERPEDMDTPRTVYKVDPSTPGSDVAAETAAALAAASLVFRKSDPAYSSRLVARAK 139
>sptr|O22297|O22297 Acidic cellulase.
Length = 505
Score = 60.8 bits (146), Expect = 4e-09
Identities = 42/106 (39%), Positives = 55/106 (51%), Gaps = 5/106 (4%)
Frame = -3
Query: 335 LDHIKELIKWGTDYILKTFNSSADTIDRIVAQVGVGDTSKGSSQPN-DHYCWMRPEDIDY 159
L++ K I+WGTDY+LK +S T + QVG PN DH+CW RPED+D
Sbjct: 123 LENAKAAIRWGTDYLLK---ASTATPGALYVQVG---------DPNMDHHCWERPEDMDT 170
Query: 158 KR---PVTECHSCSDXXXXXXXXXXXXSIVFKDS-KTYSDKLVKGA 33
R V+ + SD S+VFKDS +YS KL+K A
Sbjct: 171 PRNVYKVSTQNPGSDVAAETAAALAAASVVFKDSDPSYSTKLLKTA 216
>sptr|Q9S947|Q9S947 CEL5=CELLULASE 5 (Fragment).
Length = 169
Score = 60.1 bits (144), Expect = 7e-09
Identities = 39/107 (36%), Positives = 57/107 (53%), Gaps = 4/107 (3%)
Frame = -3
Query: 338 ELDHIKELIKWGTDYILKTFNSSADTIDRIVAQVGVGDTSKGSSQPNDHYCWMRPEDIDY 159
+L++ K I+W +DY+LK +S+DT+ V VGD ++ DH CW RPED+D
Sbjct: 36 QLNNAKAAIRWSSDYLLKAATASSDTL-----YVQVGDPNQ------DHRCWERPEDMDT 84
Query: 158 KR---PVTECHSCSDXXXXXXXXXXXXSIVFKDS-KTYSDKLVKGAK 30
R V+ + SD SIVFKDS +YS L++ A+
Sbjct: 85 PRNVYKVSPQNPGSDVAAETAAALAAASIVFKDSDPSYSSSLLRTAQ 131
>sptr|Q9LR07|Q9LR07 F10A5.13 (Hypothetical protein) (Putative
endo-beta-1,4-glucanase).
Length = 525
Score = 59.7 bits (143), Expect = 9e-09
Identities = 43/124 (34%), Positives = 59/124 (47%), Gaps = 8/124 (6%)
Frame = -3
Query: 371 AWSRPRY----EAIGELDHIKELIKWGTDYILKTFNSSADTIDRIVAQVGVGDTSKGSSQ 204
+WS Y A+ +LD K+ ++W TDY++K S V + VGD
Sbjct: 125 SWSILEYGDQMNAVNQLDPAKDSLRWITDYLIKAHPSDN------VLYIQVGDPKV---- 174
Query: 203 PNDHYCWMRPEDIDYKRPVTECH---SCSDXXXXXXXXXXXXSIVFKDS-KTYSDKLVKG 36
DH CW RPED+ KRP+T+ ++ S+VFKDS TYS L+K
Sbjct: 175 --DHPCWERPEDMKEKRPLTKIDVDTPGTEVAAETAAAMASASLVFKDSDPTYSATLLKH 232
Query: 35 AKAL 24
AK L
Sbjct: 233 AKQL 236
>sptr|Q8LCP6|Q8LCP6 Endo-beta-1,4-glucanase, putative.
Length = 525
Score = 59.7 bits (143), Expect = 9e-09
Identities = 43/124 (34%), Positives = 59/124 (47%), Gaps = 8/124 (6%)
Frame = -3
Query: 371 AWSRPRY----EAIGELDHIKELIKWGTDYILKTFNSSADTIDRIVAQVGVGDTSKGSSQ 204
+WS Y A+ +LD K+ ++W TDY++K S V + VGD
Sbjct: 125 SWSILEYGDQMNAVNQLDPAKDSLRWITDYLIKAHPSDN------VLYIQVGDPKV---- 174
Query: 203 PNDHYCWMRPEDIDYKRPVTECH---SCSDXXXXXXXXXXXXSIVFKDS-KTYSDKLVKG 36
DH CW RPED+ KRP+T+ ++ S+VFKDS TYS L+K
Sbjct: 175 --DHPCWERPEDMKEKRPLTKIDVDTPGTEVAAETAAAMASASLVFKDSDPTYSATLLKH 232
Query: 35 AKAL 24
AK L
Sbjct: 233 AKQL 236
>sptr|Q9SUS0|Q9SUS0 Putative cellulase.
Length = 479
Score = 59.7 bits (143), Expect = 9e-09
Identities = 39/124 (31%), Positives = 59/124 (47%), Gaps = 8/124 (6%)
Frame = -3
Query: 371 AWSRPRYE----AIGELDHIKELIKWGTDYILKTFNSSADTIDRIVAQVGVGDTSKGSSQ 204
+W+ Y+ ++ +L +++ IKWGTD+IL+ S + + QVG G++
Sbjct: 92 SWAAIEYQNEISSVNQLGYLRSTIKWGTDFILRAHTSP----NMLYTQVGDGNS------ 141
Query: 203 PNDHYCWMRPEDIDYKRPVTECHSC---SDXXXXXXXXXXXXSIVFKD-SKTYSDKLVKG 36
DH CW RPED+D R + S S+ S+VFK TYS L+
Sbjct: 142 --DHSCWERPEDMDTSRTLYSISSSSPGSEAAGEAAAALAAASLVFKSVDSTYSSTLLNH 199
Query: 35 AKAL 24
AK L
Sbjct: 200 AKTL 203
>sptr|Q9SZ90|Q9SZ90 Cellulase-like protein.
Length = 480
Score = 59.7 bits (143), Expect = 9e-09
Identities = 39/125 (31%), Positives = 62/125 (49%), Gaps = 8/125 (6%)
Frame = -3
Query: 371 AWSRPRYEA----IGELDHIKELIKWGTDYILKTFNSSADTIDRIVAQVGVGDTSKGSSQ 204
+W+ Y+ + +L +++ IKWGT++IL+ S+ + + QVG G++
Sbjct: 92 SWAALEYQNEITFVNQLGYLRSTIKWGTNFILRAHTST----NMLYTQVGDGNS------ 141
Query: 203 PNDHYCWMRPEDIDYKRPVTECHSC---SDXXXXXXXXXXXXSIVFK-DSKTYSDKLVKG 36
DH CW RPED+D R + S S+ S+VFK TYS KL+
Sbjct: 142 --DHSCWERPEDMDTPRTLYSISSSSPGSEAAGEAAAALAAASLVFKLVDSTYSSKLLNN 199
Query: 35 AKALS 21
AK++S
Sbjct: 200 AKSVS 204
Database: /db/uniprot/tmp/swall
Posted date: Mar 5, 2004 7:45 PM
Number of letters in database: 442,889,342
Number of sequences in database: 1,395,590
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 269,804,897
Number of Sequences: 1395590
Number of extensions: 4719802
Number of successful extensions: 10415
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 10146
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 10270
length of database: 442,889,342
effective HSP length: 99
effective length of database: 304,725,932
effective search space used: 7313422368
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

