BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= 3802297.2.1
(606 letters)
Database: /db/uniprot/tmp/swall
1,395,590 sequences; 442,889,342 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
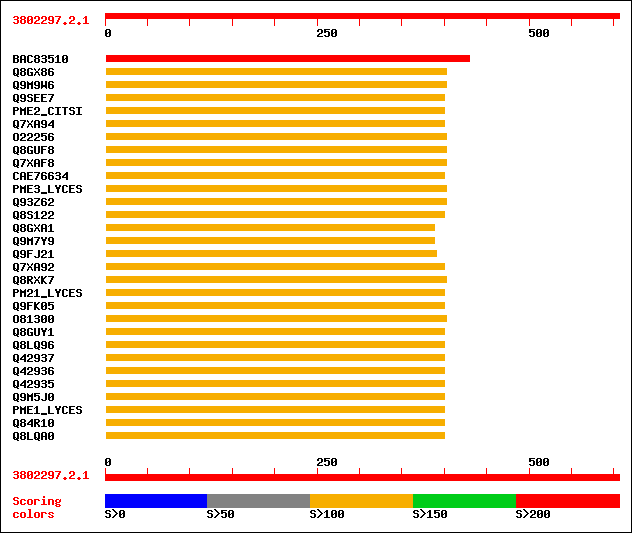
sptrnew|BAC83510|BAC83510 Putative pectin methylesterase. 215 3e-55
sptr|Q8GX86|Q8GX86 Putative pectinesterase (At3g05610). 137 1e-31
sptr|Q9M9W6|Q9M9W6 Putative pectinesterase. 137 1e-31
sptr|Q9SEE7|Q9SEE7 Pectin methyl esterase (EC 3.1.1.11) (Pectine... 134 9e-31
sw|O04887|PME2_CITSI Pectinesterase 2.1 precursor (EC 3.1.1.11) ... 134 9e-31
sptr|Q7XA94|Q7XA94 Pectinesterase (EC 3.1.1.11) (Fragment). 133 2e-30
sptr|O22256|O22256 Putative pectinesterase. 132 3e-30
sptr|Q8GUF8|Q8GUF8 At2g47550/T30B22.15. 132 3e-30
sptr|Q7XAF8|Q7XAF8 Papillar cell-specific pectin methylesterase-... 132 3e-30
sptrnew|CAE76634|CAE76634 Pectin methylesterase (EC 3.1.1.11) (F... 131 6e-30
sw|Q96576|PME3_LYCES Pectinesterase 3 precursor (EC 3.1.1.11) (P... 131 6e-30
sptr|Q93Z62|Q93Z62 At2g47550/T30B22.15. 131 8e-30
sptr|Q8S122|Q8S122 Putative pectin esterase. 131 8e-30
sptr|Q8GXA1|Q8GXA1 Putative pectin methylesterase. 130 1e-29
sptr|Q9M7Y9|Q9M7Y9 Pectin methylesterase, putative (EC 3.1.1.11)... 130 1e-29
sptr|Q9FJ21|Q9FJ21 Pectin methylesterase (EC 3.1.1.11) (AT5g4918... 129 2e-29
sptr|Q7XA92|Q7XA92 Pectinesterase (EC 3.1.1.11) (Fragment). 128 5e-29
sptr|Q8RXK7|Q8RXK7 Hypothetical protein. 128 5e-29
sw|P09607|PM21_LYCES Pectinesterase 2 precursor (EC 3.1.1.11) (P... 127 1e-28
sptr|Q9FK05|Q9FK05 Pectinesterase. 127 1e-28
sptr|O81300|O81300 T14P8.14 protein (AT4G02330 protein) (Hypothe... 127 1e-28
sptr|Q8GUY1|Q8GUY1 Methyl pectinesterase (Fragment). 127 1e-28
sptr|Q8LQ96|Q8LQ96 Putative pectinesterase 2.1. 127 1e-28
sptr|Q42937|Q42937 Pectin methylesterase (EC 3.1.1.11) (Fragment). 126 2e-28
sptr|Q42936|Q42936 Pectin methylesterase (EC 3.1.1.11) (Pectines... 126 2e-28
sptr|Q42935|Q42935 Pectin methylesterase (EC 3.1.1.11) (Pectines... 126 2e-28
sptr|Q9M5J0|Q9M5J0 Pectin methylesterase isoform alpha (EC 3.1.1... 126 2e-28
sw|P14280|PME1_LYCES Pectinesterase 1 precursor (EC 3.1.1.11) (P... 126 2e-28
sptr|Q84R10|Q84R10 Putative pectinesterase. 124 7e-28
sptr|Q8LQA0|Q8LQA0 Putative pectinesterase. 124 7e-28
>sptrnew|BAC83510|BAC83510 Putative pectin methylesterase.
Length = 566
Score = 215 bits (548), Expect = 3e-55
Identities = 89/143 (62%), Positives = 120/143 (83%)
Frame = +2
Query: 2 AQGRKERRSVGGTVIHNCTVAPHPEFEKSVGRFRTFLGRPWKEHSRTLYIQSEIGGFIDP 181
AQGR+E+RS GGTVIHNCTVAPHP+ EK + +T+L RPWKE+SRT+++Q+EIG +DP
Sbjct: 424 AQGRREKRSAGGTVIHNCTVAPHPDLEKFTDKVKTYLARPWKEYSRTIFVQNEIGAVVDP 483
Query: 182 QGWLPWLGDFGLSTCYYAKVENHGPGANMSERVKWRGIKNITYQHAMQKYTVESFIQGQH 361
GWL W G+F L T YYA+V+NHGPGA+MS+R KW+G++++TYQ +++TVE+FIQGQ
Sbjct: 484 VGWLEWNGNFALDTLYYAEVDNHGPGADMSKRAKWKGVQSLTYQDVQKEFTVEAFIQGQE 543
Query: 362 WLPQLGVPFIPGLLPQQQSGRIH 430
++P+ GVP+IPGLLPQ Q GR+H
Sbjct: 544 FIPKFGVPYIPGLLPQTQQGRMH 566
>sptr|Q8GX86|Q8GX86 Putative pectinesterase (At3g05610).
Length = 669
Score = 137 bits (344), Expect = 1e-31
Identities = 61/134 (45%), Positives = 87/134 (64%)
Frame = +2
Query: 2 AQGRKERRSVGGTVIHNCTVAPHPEFEKSVGRFRTFLGRPWKEHSRTLYIQSEIGGFIDP 181
A GRK+ R G V CT+A P++ + +LGRPWKE+SRT+ + + I F+ P
Sbjct: 434 AHGRKDPRESTGFVFQGCTIAGEPDYLAVKETSKAYLGRPWKEYSRTIIMNTFIPDFVQP 493
Query: 182 QGWLPWLGDFGLSTCYYAKVENHGPGANMSERVKWRGIKNITYQHAMQKYTVESFIQGQH 361
QGW PWLGDFGL T +Y++V+N GPG+ ++ RV W GIK ++ + + K+T +IQG
Sbjct: 494 QGWQPWLGDFGLKTLFYSEVQNTGPGSALANRVTWAGIKTLSEEDIL-KFTPAQYIQGDD 552
Query: 362 WLPQLGVPFIPGLL 403
W+P GVP+ GLL
Sbjct: 553 WIPGKGVPYTTGLL 566
>sptr|Q9M9W6|Q9M9W6 Putative pectinesterase.
Length = 669
Score = 137 bits (344), Expect = 1e-31
Identities = 61/134 (45%), Positives = 87/134 (64%)
Frame = +2
Query: 2 AQGRKERRSVGGTVIHNCTVAPHPEFEKSVGRFRTFLGRPWKEHSRTLYIQSEIGGFIDP 181
A GRK+ R G V CT+A P++ + +LGRPWKE+SRT+ + + I F+ P
Sbjct: 434 AHGRKDPRESTGFVFQGCTIAGEPDYLAVKETSKAYLGRPWKEYSRTIIMNTFIPDFVQP 493
Query: 182 QGWLPWLGDFGLSTCYYAKVENHGPGANMSERVKWRGIKNITYQHAMQKYTVESFIQGQH 361
QGW PWLGDFGL T +Y++V+N GPG+ ++ RV W GIK ++ + + K+T +IQG
Sbjct: 494 QGWQPWLGDFGLKTLFYSEVQNTGPGSALANRVTWAGIKTLSEEDIL-KFTPAQYIQGDD 552
Query: 362 WLPQLGVPFIPGLL 403
W+P GVP+ GLL
Sbjct: 553 WIPGKGVPYTTGLL 566
>sptr|Q9SEE7|Q9SEE7 Pectin methyl esterase (EC 3.1.1.11)
(Pectinesterase) (Pectin methylesterase) (Fragment).
Length = 530
Score = 134 bits (337), Expect = 9e-31
Identities = 63/133 (47%), Positives = 79/133 (59%)
Frame = +2
Query: 2 AQGRKERRSVGGTVIHNCTVAPHPEFEKSVGRFRTFLGRPWKEHSRTLYIQSEIGGFIDP 181
AQGR + GT I C + P+ E V F+T+LGRPWKE+SRT+ +QS +GG IDP
Sbjct: 396 AQGRTDPNQATGTSIQFCDIIASPDLEPVVKEFKTYLGRPWKEYSRTVVMQSYLGGLIDP 455
Query: 182 QGWLPWLGDFGLSTCYYAKVENHGPGANMSERVKWRGIKNITYQHAMQKYTVESFIQGQH 361
GW W G+F L T YY + N+GPGA S+RVKW G IT +TV IQG
Sbjct: 456 AGWAEWSGEFALKTLYYGEYMNNGPGAGTSKRVKWPGYHVITDPAEAMPFTVAELIQGGS 515
Query: 362 WLPQLGVPFIPGL 400
WL GV ++ GL
Sbjct: 516 WLSSTGVAYVDGL 528
>sw|O04887|PME2_CITSI Pectinesterase 2.1 precursor (EC 3.1.1.11)
(Pectin methylesterase) (PE).
Length = 510
Score = 134 bits (337), Expect = 9e-31
Identities = 60/133 (45%), Positives = 83/133 (62%)
Frame = +2
Query: 2 AQGRKERRSVGGTVIHNCTVAPHPEFEKSVGRFRTFLGRPWKEHSRTLYIQSEIGGFIDP 181
AQGR + G +IHNC V + + +TFLGRPWK++SRT+YI++ + I+P
Sbjct: 378 AQGRTDPNQSTGIIIHNCRVTAASDLKPVQSSVKTFLGRPWKQYSRTVYIKTFLDSLINP 437
Query: 182 QGWLPWLGDFGLSTCYYAKVENHGPGANMSERVKWRGIKNITYQHAMQKYTVESFIQGQH 361
GW+ W GDF L+T YYA+ N GPG++ + RVKWRG +T + ++TV +FI G
Sbjct: 438 AGWMEWSGDFALNTLYYAEYMNTGPGSSTANRVKWRGYHVLTSPSQVSQFTVGNFIAGNS 497
Query: 362 WLPQLGVPFIPGL 400
WLP VPF GL
Sbjct: 498 WLPATNVPFTSGL 510
>sptr|Q7XA94|Q7XA94 Pectinesterase (EC 3.1.1.11) (Fragment).
Length = 193
Score = 133 bits (334), Expect = 2e-30
Identities = 61/133 (45%), Positives = 79/133 (59%)
Frame = +2
Query: 2 AQGRKERRSVGGTVIHNCTVAPHPEFEKSVGRFRTFLGRPWKEHSRTLYIQSEIGGFIDP 181
AQ R++ G +IHNC + P+ G FRT+LGRPW+++SR + ++S + G I P
Sbjct: 61 AQSRRDPNENTGIIIHNCRITAAPDLRAVQGSFRTYLGRPWQKYSRVVIMKSNLDGLIAP 120
Query: 182 QGWLPWLGDFGLSTCYYAKVENHGPGANMSERVKWRGIKNITYQHAMQKYTVESFIQGQH 361
QGW PW G FGL T YY + N G GA RVKW G + IT K+TV +F+ G
Sbjct: 121 QGWFPWSGSFGLDTLYYGEYMNTGAGAATGGRVKWPGFRVITSATEAGKFTVGNFLAGDA 180
Query: 362 WLPQLGVPFIPGL 400
WLP GVPF GL
Sbjct: 181 WLPGTGVPFEAGL 193
>sptr|O22256|O22256 Putative pectinesterase.
Length = 560
Score = 132 bits (333), Expect = 3e-30
Identities = 62/134 (46%), Positives = 84/134 (62%)
Frame = +2
Query: 2 AQGRKERRSVGGTVIHNCTVAPHPEFEKSVGRFRTFLGRPWKEHSRTLYIQSEIGGFIDP 181
AQGR + GT IH CT+ P + S +T+LGRPWKE+SRT+ +Q+ I GF++P
Sbjct: 427 AQGRTDPNQNTGTAIHGCTIRPADDLATSNYTVKTYLGRPWKEYSRTVVMQTYIDGFLEP 486
Query: 182 QGWLPWLGDFGLSTCYYAKVENHGPGANMSERVKWRGIKNITYQHAMQKYTVESFIQGQH 361
GW W GDF LST YYA+ N GPG++ + RV W G I A +TV +F+ G+
Sbjct: 487 SGWNAWSGDFALSTLYYAEYNNTGPGSDTTNRVTWPGYHVINATDA-SNFTVTNFLVGEG 545
Query: 362 WLPQLGVPFIPGLL 403
W+ Q GVPF+ GL+
Sbjct: 546 WIGQTGVPFVGGLI 559
>sptr|Q8GUF8|Q8GUF8 At2g47550/T30B22.15.
Length = 345
Score = 132 bits (333), Expect = 3e-30
Identities = 62/134 (46%), Positives = 84/134 (62%)
Frame = +2
Query: 2 AQGRKERRSVGGTVIHNCTVAPHPEFEKSVGRFRTFLGRPWKEHSRTLYIQSEIGGFIDP 181
AQGR + GT IH CT+ P + S +T+LGRPWKE+SRT+ +Q+ I GF++P
Sbjct: 212 AQGRTDPNQNTGTAIHGCTIRPADDLATSNYTVKTYLGRPWKEYSRTVVMQTYIDGFLEP 271
Query: 182 QGWLPWLGDFGLSTCYYAKVENHGPGANMSERVKWRGIKNITYQHAMQKYTVESFIQGQH 361
GW W GDF LST YYA+ N GPG++ + RV W G I A +TV +F+ G+
Sbjct: 272 SGWNAWSGDFALSTLYYAEYNNTGPGSDTTNRVTWPGYHVINATDA-SNFTVTNFLVGEG 330
Query: 362 WLPQLGVPFIPGLL 403
W+ Q GVPF+ GL+
Sbjct: 331 WIGQTGVPFVGGLI 344
>sptr|Q7XAF8|Q7XAF8 Papillar cell-specific pectin
methylesterase-like protein.
Length = 562
Score = 132 bits (332), Expect = 3e-30
Identities = 61/134 (45%), Positives = 85/134 (63%)
Frame = +2
Query: 2 AQGRKERRSVGGTVIHNCTVAPHPEFEKSVGRFRTFLGRPWKEHSRTLYIQSEIGGFIDP 181
AQGR + GTV+H CT+ P + S +T+LGRPWKE+SRT+ +Q+ I GF+DP
Sbjct: 429 AQGRTDPNQNTGTVLHGCTIRPADDLASSNYTVKTYLGRPWKEYSRTVVMQTYIDGFLDP 488
Query: 182 QGWLPWLGDFGLSTCYYAKVENHGPGANMSERVKWRGIKNITYQHAMQKYTVESFIQGQH 361
GW W G+F LST YYA+ N GPG++ + RV W G I A +TV +F+ G+
Sbjct: 489 TGWNAWSGNFALSTLYYAEYNNTGPGSSTTNRVTWPGYHVINATDA-SNFTVTNFLVGEG 547
Query: 362 WLPQLGVPFIPGLL 403
W+ Q GVPF+ G++
Sbjct: 548 WIGQTGVPFVGGMI 561
>sptrnew|CAE76634|CAE76634 Pectin methylesterase (EC 3.1.1.11)
(Fragment).
Length = 254
Score = 131 bits (330), Expect = 6e-30
Identities = 60/133 (45%), Positives = 79/133 (59%)
Frame = +2
Query: 2 AQGRKERRSVGGTVIHNCTVAPHPEFEKSVGRFRTFLGRPWKEHSRTLYIQSEIGGFIDP 181
AQ RK+ G IHNC + P+ E S G F+T+LGRPWK +S+T+Y+ S +G I P
Sbjct: 120 AQNRKDPNQNTGISIHNCRILATPDLEASKGSFQTYLGRPWKLYSKTVYMLSYMGDHIHP 179
Query: 182 QGWLPWLGDFGLSTCYYAKVENHGPGANMSERVKWRGIKNITYQHAMQKYTVESFIQGQH 361
+GWL W F L T YY + N+GPG + +RV W G + IT ++TV FI G
Sbjct: 180 RGWLEWNATFALDTLYYGEYMNYGPGGAIGQRVTWPGFRVITSTVEANRFTVAQFISGSS 239
Query: 362 WLPQLGVPFIPGL 400
WLP GV F+ GL
Sbjct: 240 WLPSTGVAFVAGL 252
>sw|Q96576|PME3_LYCES Pectinesterase 3 precursor (EC 3.1.1.11)
(Pectin methylesterase 3) (PE 3).
Length = 544
Score = 131 bits (330), Expect = 6e-30
Identities = 60/134 (44%), Positives = 79/134 (58%)
Frame = +2
Query: 2 AQGRKERRSVGGTVIHNCTVAPHPEFEKSVGRFRTFLGRPWKEHSRTLYIQSEIGGFIDP 181
AQGR + GT I C + P+ E + ++T+LGRPWK+HSRT+ +QS + G IDP
Sbjct: 410 AQGRTDPNQATGTSIQFCDIIASPDLEPVMNEYKTYLGRPWKKHSRTVVMQSYLDGHIDP 469
Query: 182 QGWLPWLGDFGLSTCYYAKVENHGPGANMSERVKWRGIKNITYQHAMQKYTVESFIQGQH 361
GW W GDF L T YY + N+GPGA S+RVKW G IT + +TV IQG
Sbjct: 470 SGWFEWRGDFALKTLYYGEFMNNGPGAGTSKRVKWPGYHVITDPNEAMPFTVAELIQGGS 529
Query: 362 WLPQLGVPFIPGLL 403
WL V ++ GL+
Sbjct: 530 WLNSTSVAYVEGLV 543
>sptr|Q93Z62|Q93Z62 At2g47550/T30B22.15.
Length = 345
Score = 131 bits (329), Expect = 8e-30
Identities = 62/134 (46%), Positives = 83/134 (61%)
Frame = +2
Query: 2 AQGRKERRSVGGTVIHNCTVAPHPEFEKSVGRFRTFLGRPWKEHSRTLYIQSEIGGFIDP 181
AQGR GT IH CT+ P + S +T+LGRPWKE+SRT+ +Q+ I GF++P
Sbjct: 212 AQGRTYPNQNTGTAIHGCTIRPADDLATSNYTVKTYLGRPWKEYSRTVVMQTYIDGFLEP 271
Query: 182 QGWLPWLGDFGLSTCYYAKVENHGPGANMSERVKWRGIKNITYQHAMQKYTVESFIQGQH 361
GW W GDF LST YYA+ N GPG++ + RV W G I A +TV +F+ G+
Sbjct: 272 SGWNAWSGDFALSTLYYAEYNNTGPGSDTTNRVTWPGYHVINATDA-SNFTVTNFLVGEG 330
Query: 362 WLPQLGVPFIPGLL 403
W+ Q GVPF+ GL+
Sbjct: 331 WIGQTGVPFVGGLI 344
>sptr|Q8S122|Q8S122 Putative pectin esterase.
Length = 563
Score = 131 bits (329), Expect = 8e-30
Identities = 63/134 (47%), Positives = 82/134 (61%), Gaps = 1/134 (0%)
Frame = +2
Query: 2 AQGRKERRSVGGTVIHNCTVAPHPEFEKSVGRFRTFLGRPWKEHSRTLYIQSEIGGFIDP 181
AQGR + GT IH C V P P+ + +F TFLGRPWKE+SRT+Y+ S + +DP
Sbjct: 430 AQGRTDPNQNTGTSIHRCRVVPAPDLAPAAKQFPTFLGRPWKEYSRTVYMLSYLDSHVDP 489
Query: 182 QGWLPWLG-DFGLSTCYYAKVENHGPGANMSERVKWRGIKNITYQHAMQKYTVESFIQGQ 358
+GWL W G DF L T +Y + +N GPGA+ + RV W G IT Q ++TV FIQG
Sbjct: 490 RGWLEWNGADFALKTLFYGEYQNQGPGASTAGRVNWPGYHVITDQSVAMQFTVGQFIQGG 549
Query: 359 HWLPQLGVPFIPGL 400
+WL GV + GL
Sbjct: 550 NWLKATGVNYNEGL 563
>sptr|Q8GXA1|Q8GXA1 Putative pectin methylesterase.
Length = 568
Score = 130 bits (328), Expect = 1e-29
Identities = 61/129 (47%), Positives = 81/129 (62%)
Frame = +2
Query: 2 AQGRKERRSVGGTVIHNCTVAPHPEFEKSVGRFRTFLGRPWKEHSRTLYIQSEIGGFIDP 181
AQGR R G V+H C + P + + +LGRPWKE SRT+ +++ I IDP
Sbjct: 437 AQGRSNVRESTGLVLHGCHITGDPAYIPMKSVNKAYLGRPWKEFSRTIIMKTTIDDVIDP 496
Query: 182 QGWLPWLGDFGLSTCYYAKVENHGPGANMSERVKWRGIKNITYQHAMQKYTVESFIQGQH 361
GWLPW GDF L T YYA+ N GPG+N ++RVKW GIK +T Q A+ YT + F++G
Sbjct: 497 AGWLPWSGDFALKTLYYAEHMNTGPGSNQAQRVKWPGIKKLTPQDAL-LYTGDRFLRGDT 555
Query: 362 WLPQLGVPF 388
W+PQ VP+
Sbjct: 556 WIPQTQVPY 564
>sptr|Q9M7Y9|Q9M7Y9 Pectin methylesterase, putative (EC 3.1.1.11)
(Pectinesterase).
Length = 562
Score = 130 bits (328), Expect = 1e-29
Identities = 61/129 (47%), Positives = 81/129 (62%)
Frame = +2
Query: 2 AQGRKERRSVGGTVIHNCTVAPHPEFEKSVGRFRTFLGRPWKEHSRTLYIQSEIGGFIDP 181
AQGR R G V+H C + P + + +LGRPWKE SRT+ +++ I IDP
Sbjct: 431 AQGRSNVRESTGLVLHGCHITGDPAYIPMKSVNKAYLGRPWKEFSRTIIMKTTIDDVIDP 490
Query: 182 QGWLPWLGDFGLSTCYYAKVENHGPGANMSERVKWRGIKNITYQHAMQKYTVESFIQGQH 361
GWLPW GDF L T YYA+ N GPG+N ++RVKW GIK +T Q A+ YT + F++G
Sbjct: 491 AGWLPWSGDFALKTLYYAEHMNTGPGSNQAQRVKWPGIKKLTPQDAL-LYTGDRFLRGDT 549
Query: 362 WLPQLGVPF 388
W+PQ VP+
Sbjct: 550 WIPQTQVPY 558
>sptr|Q9FJ21|Q9FJ21 Pectin methylesterase (EC 3.1.1.11)
(AT5g49180/K21P3_5) (Pectinesterase).
Length = 571
Score = 129 bits (325), Expect = 2e-29
Identities = 58/130 (44%), Positives = 85/130 (65%)
Frame = +2
Query: 2 AQGRKERRSVGGTVIHNCTVAPHPEFEKSVGRFRTFLGRPWKEHSRTLYIQSEIGGFIDP 181
AQGR ++R G V+ NC + P + + +LGRPWKE SRT+ + + I IDP
Sbjct: 439 AQGRSDKRESTGLVLQNCHITGEPAYIPVKSINKAYLGRPWKEFSRTIIMGTTIDDVIDP 498
Query: 182 QGWLPWLGDFGLSTCYYAKVENHGPGANMSERVKWRGIKNITYQHAMQKYTVESFIQGQH 361
GWLPW GDF L+T YYA+ EN+GPG+N ++RVKW GIK ++ + A+ ++T F++G
Sbjct: 499 AGWLPWNGDFALNTLYYAEYENNGPGSNQAQRVKWPGIKKLSPKQAL-RFTPARFLRGNL 557
Query: 362 WLPQLGVPFI 391
W+P VP++
Sbjct: 558 WIPPNRVPYM 567
>sptr|Q7XA92|Q7XA92 Pectinesterase (EC 3.1.1.11) (Fragment).
Length = 211
Score = 128 bits (322), Expect = 5e-29
Identities = 61/133 (45%), Positives = 76/133 (57%)
Frame = +2
Query: 2 AQGRKERRSVGGTVIHNCTVAPHPEFEKSVGRFRTFLGRPWKEHSRTLYIQSEIGGFIDP 181
AQ RK+ G IH C + P+ E G F TFLGRPWK +SR +Y+ S IG + P
Sbjct: 77 AQNRKDPNQNTGMSIHACRILATPDLEPVKGSFPTFLGRPWKMYSRVVYMLSYIGDHVQP 136
Query: 182 QGWLPWLGDFGLSTCYYAKVENHGPGANMSERVKWRGIKNITYQHAMQKYTVESFIQGQH 361
+GWL W F L T YY + N+GPG + +RVKW G + IT K+TV FI G
Sbjct: 137 RGWLEWNTTFALDTLYYGEYMNYGPGGAIGQRVKWPGYRVITSMVEASKFTVAEFIYGSS 196
Query: 362 WLPQLGVPFIPGL 400
WLP GV F+ GL
Sbjct: 197 WLPSTGVAFLGGL 209
>sptr|Q8RXK7|Q8RXK7 Hypothetical protein.
Length = 573
Score = 128 bits (322), Expect = 5e-29
Identities = 62/134 (46%), Positives = 84/134 (62%)
Frame = +2
Query: 2 AQGRKERRSVGGTVIHNCTVAPHPEFEKSVGRFRTFLGRPWKEHSRTLYIQSEIGGFIDP 181
AQGR ++ G IHNCT+ P + S +T+LGRPWKE+SRT+++QS I ++P
Sbjct: 440 AQGRTDQNQNTGISIHNCTIKPADDLVSSNYTVKTYLGRPWKEYSRTVFMQSYIDEVVEP 499
Query: 182 QGWLPWLGDFGLSTCYYAKVENHGPGANMSERVKWRGIKNITYQHAMQKYTVESFIQGQH 361
GW W GDF LST YYA+ N G G++ ++RV W G I A +TVE+F+ G
Sbjct: 500 VGWREWNGDFALSTLYYAEYNNTGSGSSTTDRVVWPGYHVINSTDA-NNFTVENFLLGDG 558
Query: 362 WLPQLGVPFIPGLL 403
W+ Q GVP+I GLL
Sbjct: 559 WMVQSGVPYISGLL 572
>sw|P09607|PM21_LYCES Pectinesterase 2 precursor (EC 3.1.1.11)
(Pectin methylesterase 2) (PE 2).
Length = 550
Score = 127 bits (319), Expect = 1e-28
Identities = 60/133 (45%), Positives = 77/133 (57%)
Frame = +2
Query: 2 AQGRKERRSVGGTVIHNCTVAPHPEFEKSVGRFRTFLGRPWKEHSRTLYIQSEIGGFIDP 181
AQGR + GT I C + P+ + V F T+LGRPWK++SRT+ ++S +GG IDP
Sbjct: 416 AQGRTDPNQATGTSIQFCDIIASPDLKPVVKEFPTYLGRPWKKYSRTVVMESYLGGLIDP 475
Query: 182 QGWLPWLGDFGLSTCYYAKVENHGPGANMSERVKWRGIKNITYQHAMQKYTVESFIQGQH 361
GW W GDF L T YY + N+GPGA S+RVKW G IT +TV IQG
Sbjct: 476 SGWAEWHGDFALKTLYYGEFMNNGPGAGTSKRVKWPGYHVITDPAEAMSFTVAKLIQGGS 535
Query: 362 WLPQLGVPFIPGL 400
WL V ++ GL
Sbjct: 536 WLRSTDVAYVDGL 548
>sptr|Q9FK05|Q9FK05 Pectinesterase.
Length = 587
Score = 127 bits (319), Expect = 1e-28
Identities = 60/133 (45%), Positives = 77/133 (57%)
Frame = +2
Query: 2 AQGRKERRSVGGTVIHNCTVAPHPEFEKSVGRFRTFLGRPWKEHSRTLYIQSEIGGFIDP 181
AQ RK+ G IH C + P+ E S G + T+LGRPWK +SR +Y+ S++G IDP
Sbjct: 453 AQNRKDPNQNTGISIHACKLLATPDLEASKGSYPTYLGRPWKLYSRVVYMMSDMGDHIDP 512
Query: 182 QGWLPWLGDFGLSTCYYAKVENHGPGANMSERVKWRGIKNITYQHAMQKYTVESFIQGQH 361
+GWL W G F L + YY + N G G+ + +RVKW G IT K+TV FI G
Sbjct: 513 RGWLEWNGPFALDSLYYGEYMNKGLGSGIGQRVKWPGYHVITSTVEASKFTVAQFISGSS 572
Query: 362 WLPQLGVPFIPGL 400
WLP GV F GL
Sbjct: 573 WLPSTGVSFFSGL 585
>sptr|O81300|O81300 T14P8.14 protein (AT4G02330 protein)
(Hypothetical protein).
Length = 573
Score = 127 bits (319), Expect = 1e-28
Identities = 62/134 (46%), Positives = 83/134 (61%)
Frame = +2
Query: 2 AQGRKERRSVGGTVIHNCTVAPHPEFEKSVGRFRTFLGRPWKEHSRTLYIQSEIGGFIDP 181
AQGR + G IHNCT+ P + S +T+LGRPWKE+SRT+++QS I ++P
Sbjct: 440 AQGRTDPNQNTGISIHNCTIKPADDLVSSNYTVKTYLGRPWKEYSRTVFMQSYIDEVVEP 499
Query: 182 QGWLPWLGDFGLSTCYYAKVENHGPGANMSERVKWRGIKNITYQHAMQKYTVESFIQGQH 361
GW W GDF LST YYA+ N G G++ ++RV W G I A +TVE+F+ G
Sbjct: 500 VGWREWNGDFALSTLYYAEYNNTGSGSSTTDRVVWPGYHVINSTDA-NNFTVENFLLGDG 558
Query: 362 WLPQLGVPFIPGLL 403
W+ Q GVP+I GLL
Sbjct: 559 WMVQSGVPYISGLL 572
>sptr|Q8GUY1|Q8GUY1 Methyl pectinesterase (Fragment).
Length = 226
Score = 127 bits (318), Expect = 1e-28
Identities = 60/133 (45%), Positives = 78/133 (58%)
Frame = +2
Query: 2 AQGRKERRSVGGTVIHNCTVAPHPEFEKSVGRFRTFLGRPWKEHSRTLYIQSEIGGFIDP 181
AQGR + G I C VA + FRT+LGRPWK++SRT+Y+QSE+ +DP
Sbjct: 94 AQGRTDPNQNTGISIQKCKVAAASDLAAVQTSFRTYLGRPWKQYSRTVYLQSELDSVVDP 153
Query: 182 QGWLPWLGDFGLSTCYYAKVENHGPGANMSERVKWRGIKNITYQHAMQKYTVESFIQGQH 361
+GWL W G F L T YY + +N G GA+ S RVKW+G + I+ +TV SFI G
Sbjct: 154 KGWLEWDGTFALDTLYYGEYQNTGAGASTSNRVKWKGYRVISSSSEASTFTVGSFIDGDV 213
Query: 362 WLPQLGVPFIPGL 400
WL +PF GL
Sbjct: 214 WLAGTSIPFSTGL 226
>sptr|Q8LQ96|Q8LQ96 Putative pectinesterase 2.1.
Length = 295
Score = 127 bits (318), Expect = 1e-28
Identities = 59/133 (44%), Positives = 80/133 (60%)
Frame = +2
Query: 2 AQGRKERRSVGGTVIHNCTVAPHPEFEKSVGRFRTFLGRPWKEHSRTLYIQSEIGGFIDP 181
AQGR++ G I C VA + F+T+LGRPWK++SRT+++QSE+ ++P
Sbjct: 163 AQGREDPNQNTGISIQKCKVAAASDLLAVQSSFKTYLGRPWKQYSRTVFMQSELDSVVNP 222
Query: 182 QGWLPWLGDFGLSTCYYAKVENHGPGANMSERVKWRGIKNITYQHAMQKYTVESFIQGQH 361
GWL W G+F L T YY + +N GPGA+ S RVKW+G + IT +TV +FI G
Sbjct: 223 AGWLEWSGNFALDTLYYGEYQNTGPGASTSNRVKWKGYRVITSASEASTFTVGNFIDGDV 282
Query: 362 WLPQLGVPFIPGL 400
WL VPF GL
Sbjct: 283 WLAGTSVPFTVGL 295
>sptr|Q42937|Q42937 Pectin methylesterase (EC 3.1.1.11) (Fragment).
Length = 274
Score = 126 bits (317), Expect = 2e-28
Identities = 62/133 (46%), Positives = 79/133 (59%)
Frame = +2
Query: 2 AQGRKERRSVGGTVIHNCTVAPHPEFEKSVGRFRTFLGRPWKEHSRTLYIQSEIGGFIDP 181
AQGR++ GT I NC + P + G +T+LGRPWK +SRT+++QS IG IDP
Sbjct: 143 AQGREDPNQNTGTSIQNCDIIPSSDLAPVKGSVKTYLGRPWKAYSRTVFMQSNIGDHIDP 202
Query: 182 QGWLPWLGDFGLSTCYYAKVENHGPGANMSERVKWRGIKNITYQHAMQKYTVESFIQGQH 361
+GW W GDF L T YY + N GPGA S+RVKW G ++ A K+TV IQG
Sbjct: 203 EGWSVWDGDFALKTLYYGEYMNKGPGAGTSKRVKWPGYHILSAAEA-TKFTVGQLIQGGV 261
Query: 362 WLPQLGVPFIPGL 400
WL GV + GL
Sbjct: 262 WLKSTGVAYTEGL 274
>sptr|Q42936|Q42936 Pectin methylesterase (EC 3.1.1.11)
(Pectinesterase) (Fragment).
Length = 315
Score = 126 bits (317), Expect = 2e-28
Identities = 62/133 (46%), Positives = 79/133 (59%)
Frame = +2
Query: 2 AQGRKERRSVGGTVIHNCTVAPHPEFEKSVGRFRTFLGRPWKEHSRTLYIQSEIGGFIDP 181
AQGR++ GT I NC + P + G +T+LGRPWK +SRT+++QS IG IDP
Sbjct: 184 AQGREDPNQNTGTSIQNCDIIPSSDLAPVKGSVKTYLGRPWKAYSRTVFMQSNIGDHIDP 243
Query: 182 QGWLPWLGDFGLSTCYYAKVENHGPGANMSERVKWRGIKNITYQHAMQKYTVESFIQGQH 361
+GW W GDF L T YY + N GPGA S+RVKW G ++ A K+TV IQG
Sbjct: 244 EGWSVWDGDFALKTLYYGEYMNKGPGAGTSKRVKWPGYHILSAAEA-TKFTVGQLIQGGV 302
Query: 362 WLPQLGVPFIPGL 400
WL GV + GL
Sbjct: 303 WLKSTGVAYTEGL 315
>sptr|Q42935|Q42935 Pectin methylesterase (EC 3.1.1.11)
(Pectinesterase) (Fragment).
Length = 315
Score = 126 bits (317), Expect = 2e-28
Identities = 62/133 (46%), Positives = 79/133 (59%)
Frame = +2
Query: 2 AQGRKERRSVGGTVIHNCTVAPHPEFEKSVGRFRTFLGRPWKEHSRTLYIQSEIGGFIDP 181
AQGR++ GT I NC + P + G +T+LGRPWK +SRT+++QS IG IDP
Sbjct: 184 AQGREDPNQNTGTSIQNCDIIPSSDLAPVKGSVKTYLGRPWKAYSRTVFMQSNIGDHIDP 243
Query: 182 QGWLPWLGDFGLSTCYYAKVENHGPGANMSERVKWRGIKNITYQHAMQKYTVESFIQGQH 361
+GW W GDF L T YY + N GPGA S+RVKW G ++ A K+TV IQG
Sbjct: 244 EGWSVWDGDFALKTLYYGEYMNKGPGAGTSKRVKWPGYHILSAAEA-TKFTVGQLIQGGV 302
Query: 362 WLPQLGVPFIPGL 400
WL GV + GL
Sbjct: 303 WLKSTGVAYTEGL 315
>sptr|Q9M5J0|Q9M5J0 Pectin methylesterase isoform alpha (EC
3.1.1.11) (Fragment).
Length = 277
Score = 126 bits (317), Expect = 2e-28
Identities = 60/133 (45%), Positives = 77/133 (57%)
Frame = +2
Query: 2 AQGRKERRSVGGTVIHNCTVAPHPEFEKSVGRFRTFLGRPWKEHSRTLYIQSEIGGFIDP 181
AQGR++ G I NC VA + FR +LGRPWK +SRT+++ S + I+P
Sbjct: 144 AQGREDPNQSTGISIINCKVAAAADLIPVKSEFRNYLGRPWKMYSRTVFLNSLMEDLIEP 203
Query: 182 QGWLPWLGDFGLSTCYYAKVENHGPGANMSERVKWRGIKNITYQHAMQKYTVESFIQGQH 361
GWL W G F L T YY + N GPGAN S RV W G + IT ++TV++FIQG
Sbjct: 204 AGWLEWNGTFALDTLYYGEYNNRGPGANTSGRVTWPGYRVITNSTEASQFTVQNFIQGNE 263
Query: 362 WLPQLGVPFIPGL 400
WL G+PF GL
Sbjct: 264 WLNSYGIPFFSGL 276
>sw|P14280|PME1_LYCES Pectinesterase 1 precursor (EC 3.1.1.11)
(Pectin methylesterase 1) (PE 1).
Length = 546
Score = 126 bits (316), Expect = 2e-28
Identities = 60/133 (45%), Positives = 77/133 (57%)
Frame = +2
Query: 2 AQGRKERRSVGGTVIHNCTVAPHPEFEKSVGRFRTFLGRPWKEHSRTLYIQSEIGGFIDP 181
AQGR + GT I C + + E + F T+LGRPWKE+SRT+ ++S +GG I+P
Sbjct: 412 AQGRTDPNQATGTSIQFCNIIASSDLEPVLKEFPTYLGRPWKEYSRTVVMESYLGGLINP 471
Query: 182 QGWLPWLGDFGLSTCYYAKVENHGPGANMSERVKWRGIKNITYQHAMQKYTVESFIQGQH 361
GW W GDF L T YY + N+GPGA S+RVKW G IT +TV IQG
Sbjct: 472 AGWAEWDGDFALKTLYYGEFMNNGPGAGTSKRVKWPGYHVITDPAKAMPFTVAKLIQGGS 531
Query: 362 WLPQLGVPFIPGL 400
WL GV ++ GL
Sbjct: 532 WLRSTGVAYVDGL 544
>sptr|Q84R10|Q84R10 Putative pectinesterase.
Length = 519
Score = 124 bits (312), Expect = 7e-28
Identities = 56/133 (42%), Positives = 79/133 (59%)
Frame = +2
Query: 2 AQGRKERRSVGGTVIHNCTVAPHPEFEKSVGRFRTFLGRPWKEHSRTLYIQSEIGGFIDP 181
AQGR + + G I + + PEFE GRF+++LGRPWK++SRT++++++I IDP
Sbjct: 387 AQGRDDPHTNSGISIQHSRIRAAPEFEAVKGRFKSYLGRPWKKYSRTVFLKTDIDELIDP 446
Query: 182 QGWLPWLGDFGLSTCYYAKVENHGPGANMSERVKWRGIKNITYQHAMQKYTVESFIQGQH 361
+GW W G + LST YY + N G GA RV W G + + +TV FIQG
Sbjct: 447 RGWREWSGSYALSTLYYGEFMNTGAGAGTGRRVNWPGFHVLRGEEEASPFTVSRFIQGDS 506
Query: 362 WLPQLGVPFIPGL 400
W+P GVPF G+
Sbjct: 507 WIPITGVPFSAGV 519
>sptr|Q8LQA0|Q8LQA0 Putative pectinesterase.
Length = 557
Score = 124 bits (312), Expect = 7e-28
Identities = 60/134 (44%), Positives = 77/134 (57%), Gaps = 1/134 (0%)
Frame = +2
Query: 2 AQGRKERRSVGGTVIHNCTVAPHPEFEKSVGRFRT-FLGRPWKEHSRTLYIQSEIGGFID 178
AQGR + GT I C + P+ + T +LGRPWK +SRT+ +QS +GG ID
Sbjct: 423 AQGRTDPNQNTGTTIQGCAIVAAPDLAANTAFATTNYLGRPWKLYSRTVIMQSVVGGLID 482
Query: 179 PQGWLPWLGDFGLSTCYYAKVENHGPGANMSERVKWRGIKNITYQHAMQKYTVESFIQGQ 358
P GW+PW GD+ LST YYA+ N G GA+ S RV W G + +TV + + G
Sbjct: 483 PAGWMPWDGDYALSTLYYAEYNNSGAGADTSRRVTWPGYHVLNSTADAGNFTVGNMVLGD 542
Query: 359 HWLPQLGVPFIPGL 400
WLPQ GVPF GL
Sbjct: 543 FWLPQTGVPFTSGL 556
Database: /db/uniprot/tmp/swall
Posted date: Mar 5, 2004 7:45 PM
Number of letters in database: 442,889,342
Number of sequences in database: 1,395,590
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 450,766,106
Number of Sequences: 1395590
Number of extensions: 10331718
Number of successful extensions: 34031
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 32719
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 33907
length of database: 442,889,342
effective HSP length: 117
effective length of database: 279,605,312
effective search space used: 23486846208
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

