BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= 3713493.2.1
(696 letters)
Database: /db/trembl-ebi/tmp/swall
1,272,877 sequences; 406,886,216 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
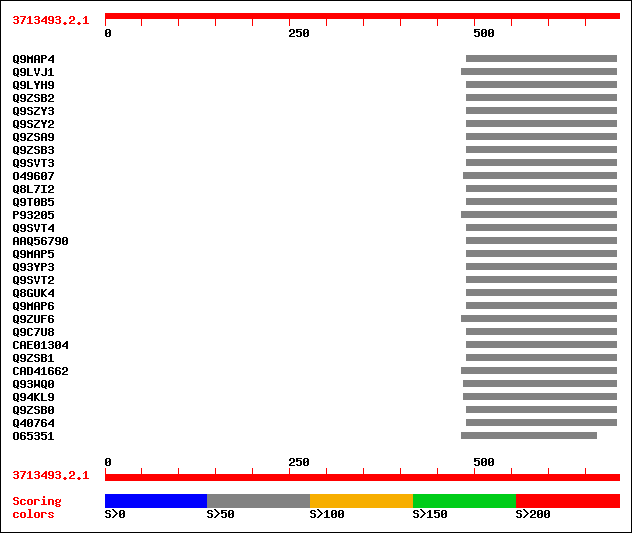
sptr|Q9MAP4|Q9MAP4 Fourth of four adjacent putative subtilase fa... 63 3e-09
sptr|Q9LVJ1|Q9LVJ1 Cucumisin-like serine protease, subtilisin-li... 62 5e-09
sptr|Q9LYH9|Q9LYH9 Subtilisin-like protease-like protein. 58 1e-07
sptr|Q9ZSB2|Q9ZSB2 F3H7.2 protein. 56 4e-07
sptr|Q9SZY3|Q9SZY3 Putative subtilisin-like protease. 56 4e-07
sptr|Q9SZY2|Q9SZY2 Putative subtilisin-like protease. 56 5e-07
sptr|Q9ZSA9|Q9ZSA9 F3H7.15 protein. 56 5e-07
sptr|Q9ZSB3|Q9ZSB3 F3H7.3 protein. 55 1e-06
sptr|Q9SVT3|Q9SVT3 Subtilisin-like protease. 55 1e-06
sptr|O49607|O49607 Subtilisin proteinase - like (Putative subtil... 55 1e-06
sptr|Q8L7I2|Q8L7I2 Putative subtilisin serine protease. 55 1e-06
sptr|Q9T0B5|Q9T0B5 Subtilisin-like protease-like protein. 55 1e-06
sptr|P93205|P93205 Serine protease, SBT2. 54 2e-06
sptr|Q9SVT4|Q9SVT4 Serine protease-like protein. 54 2e-06
sptrnew|AAQ56790|AAQ56790 At1g32960. 54 2e-06
sptr|Q9MAP5|Q9MAP5 Third of four adjacent putative subtilase fam... 54 2e-06
sptr|Q93YP3|Q93YP3 Subtilisin proteinase-like. 53 3e-06
sptr|Q9SVT2|Q9SVT2 Subtilisin proteinase-like. 53 3e-06
sptr|Q8GUK4|Q8GUK4 Subtilisin proteinase-like. 53 3e-06
sptr|Q9MAP6|Q9MAP6 Second of four adjacent putative subtilase fa... 53 3e-06
sptr|Q9ZUF6|Q9ZUF6 Putative serine protease (Putative subtilisin... 53 4e-06
sptr|Q9C7U8|Q9C7U8 Subtilisin-like protein. 53 4e-06
sptrnew|CAE01304|CAE01304 OSJNBa0020P07.21 protein. 53 4e-06
sptr|Q9ZSB1|Q9ZSB1 F3H7.13 protein (Putative subtilisin-like pro... 52 5e-06
sptrnew|CAD41662|CAD41662 OSJNBa0019K04.9 protein. 52 7e-06
sptr|Q93WQ0|Q93WQ0 Subtilisin-type protease. 52 7e-06
sptr|Q94KL9|Q94KL9 Subtilisin-like protein. 52 7e-06
sptr|Q9ZSB0|Q9ZSB0 F3H7.14 protein (Putative subtilisin-like pro... 52 9e-06
sptr|Q40764|Q40764 Antifreeze-like protein (af70). 51 1e-05
sptr|O65351|O65351 CUCUMISIN-like serine protease (Putative seri... 51 2e-05
>sptr|Q9MAP4|Q9MAP4 Fourth of four adjacent putative subtilase
family>.
Length = 734
Score = 63.2 bits (152), Expect = 3e-09
Identities = 28/68 (41%), Positives = 45/68 (66%)
Frame = -2
Query: 692 YQPTIASPYGVDVTVTPSKLAFDGKQQSLGYEITIAVSGNPVIVDSSYSFGSITWSDGAH 513
Y+P I +P GV+VTVTPS L F+ + L +++ + + IV++ Y FGS+TW+D H
Sbjct: 658 YKPVIEAPMGVNVTVTPSTLVFNAYTRKLSFKVRVLTNH---IVNTGYYFGSLTWTDSVH 714
Query: 512 DVTSPIAV 489
+V P++V
Sbjct: 715 NVVIPVSV 722
>sptr|Q9LVJ1|Q9LVJ1 Cucumisin-like serine protease, subtilisin-like
protease.
Length = 777
Score = 62.4 bits (150), Expect = 5e-09
Identities = 32/72 (44%), Positives = 43/72 (59%), Gaps = 2/72 (2%)
Frame = -2
Query: 692 YQPTIASPYGVDVTVTPSKLAFDGKQQSLGYEITI--AVSGNPVIVDSSYSFGSITWSDG 519
Y+ + SP V++ V+PSKLAF ++ L YE+T V G V + FGSI W+DG
Sbjct: 698 YEVGVKSPANVEIDVSPSKLAFSKEKSVLEYEVTFKSVVLGGGVGSVPGHEFGSIEWTDG 757
Query: 518 AHDVTSPIAVTW 483
H V SP+AV W
Sbjct: 758 EHVVKSPVAVQW 769
>sptr|Q9LYH9|Q9LYH9 Subtilisin-like protease-like protein.
Length = 755
Score = 58.2 bits (139), Expect = 1e-07
Identities = 26/68 (38%), Positives = 44/68 (64%)
Frame = -2
Query: 692 YQPTIASPYGVDVTVTPSKLAFDGKQQSLGYEITIAVSGNPVIVDSSYSFGSITWSDGAH 513
Y+ + P GV ++VTP+ L F+ + L Y++T++ + +S Y FGS+TW+DG+H
Sbjct: 681 YKLIVEPPLGVKISVTPNTLLFNSNVKILSYKVTVSTTHKS---NSIYYFGSLTWTDGSH 737
Query: 512 DVTSPIAV 489
VT P++V
Sbjct: 738 KVTIPLSV 745
>sptr|Q9ZSB2|Q9ZSB2 F3H7.2 protein.
Length = 685
Score = 56.2 bits (134), Expect = 4e-07
Identities = 24/68 (35%), Positives = 42/68 (61%)
Frame = -2
Query: 692 YQPTIASPYGVDVTVTPSKLAFDGKQQSLGYEITIAVSGNPVIVDSSYSFGSITWSDGAH 513
Y+ I P G+ VTVTP L F+ + + +++ ++ + +++ Y FGS+TWSD H
Sbjct: 609 YKVVIEPPIGIQVTVTPETLLFNSTTKRVSFKVKVSTTHK---INTGYFFGSLTWSDSLH 665
Query: 512 DVTSPIAV 489
+VT P++V
Sbjct: 666 NVTIPLSV 673
>sptr|Q9SZY3|Q9SZY3 Putative subtilisin-like protease.
Length = 775
Score = 56.2 bits (134), Expect = 4e-07
Identities = 24/68 (35%), Positives = 42/68 (61%)
Frame = -2
Query: 692 YQPTIASPYGVDVTVTPSKLAFDGKQQSLGYEITIAVSGNPVIVDSSYSFGSITWSDGAH 513
Y+ I P G+ VTVTP L F+ + + +++ ++ + +++ Y FGS+TWSD H
Sbjct: 699 YKVVIEPPIGIQVTVTPETLLFNSTTKRVSFKVKVSTTHK---INTGYFFGSLTWSDSLH 755
Query: 512 DVTSPIAV 489
+VT P++V
Sbjct: 756 NVTIPLSV 763
>sptr|Q9SZY2|Q9SZY2 Putative subtilisin-like protease.
Length = 765
Score = 55.8 bits (133), Expect = 5e-07
Identities = 23/68 (33%), Positives = 42/68 (61%)
Frame = -2
Query: 692 YQPTIASPYGVDVTVTPSKLAFDGKQQSLGYEITIAVSGNPVIVDSSYSFGSITWSDGAH 513
Y+ + P G VTVTP L F+ + + ++++++ + +++ Y FGS+TWSD H
Sbjct: 689 YRVAVEPPLGTQVTVTPETLVFNSTTKRVSFKVSVSTTHK---INTGYYFGSLTWSDSLH 745
Query: 512 DVTSPIAV 489
+VT P++V
Sbjct: 746 NVTIPLSV 753
>sptr|Q9ZSA9|Q9ZSA9 F3H7.15 protein.
Length = 774
Score = 55.8 bits (133), Expect = 5e-07
Identities = 23/68 (33%), Positives = 42/68 (61%)
Frame = -2
Query: 692 YQPTIASPYGVDVTVTPSKLAFDGKQQSLGYEITIAVSGNPVIVDSSYSFGSITWSDGAH 513
Y+ + P G VTVTP L F+ + + ++++++ + +++ Y FGS+TWSD H
Sbjct: 698 YRVAVEPPLGTQVTVTPETLVFNSTTKRVSFKVSVSTTHK---INTGYYFGSLTWSDSLH 754
Query: 512 DVTSPIAV 489
+VT P++V
Sbjct: 755 NVTIPLSV 762
>sptr|Q9ZSB3|Q9ZSB3 F3H7.3 protein.
Length = 751
Score = 54.7 bits (130), Expect = 1e-06
Identities = 24/68 (35%), Positives = 41/68 (60%)
Frame = -2
Query: 692 YQPTIASPYGVDVTVTPSKLAFDGKQQSLGYEITIAVSGNPVIVDSSYSFGSITWSDGAH 513
Y+ T+ P G VTVTP L F+ + + +++ ++ + ++ Y FGS+TWSD H
Sbjct: 675 YRVTVEPPLGFQVTVTPETLVFNSTTKKVYFKVKVSTTHK---TNTGYYFGSLTWSDSLH 731
Query: 512 DVTSPIAV 489
+VT P++V
Sbjct: 732 NVTIPLSV 739
>sptr|Q9SVT3|Q9SVT3 Subtilisin-like protease.
Length = 769
Score = 54.7 bits (130), Expect = 1e-06
Identities = 28/69 (40%), Positives = 41/69 (59%), Gaps = 1/69 (1%)
Frame = -2
Query: 692 YQPTIASPYGVDVTVTPSKLAF-DGKQQSLGYEITIAVSGNPVIVDSSYSFGSITWSDGA 516
Y+ I SP G+ +TV P+ L F ++ L + + S V+S Y FGS+TW+DG
Sbjct: 698 YRAVIESPLGITLTVNPTILVFKSAAKRVLTFSVKAKTSHK---VNSGYFFGSLTWTDGV 754
Query: 515 HDVTSPIAV 489
HDVT P++V
Sbjct: 755 HDVTIPVSV 763
>sptr|O49607|O49607 Subtilisin proteinase - like (Putative
subtilisin proteinase) (Putative subtilisin serine
proteinase).
Length = 764
Score = 54.7 bits (130), Expect = 1e-06
Identities = 31/71 (43%), Positives = 41/71 (57%), Gaps = 2/71 (2%)
Frame = -2
Query: 692 YQPTIASPYGVDVTVTPSKLAFDGKQQSLGYEITIAVSG-NPVIVDSSYSFGSITWSDGA 516
Y+ I SP GV VTV P +L F + Y +T+ V+ N V+ ++ FGS+TW DG
Sbjct: 689 YRARIESPRGVTVTVKPPRLVFTSAVKRRSYAVTVTVNTRNVVLGETGAVFGSVTWFDGG 748
Query: 515 -HDVTSPIAVT 486
H V SPI VT
Sbjct: 749 KHVVRSPIVVT 759
>sptr|Q8L7I2|Q8L7I2 Putative subtilisin serine protease.
Length = 778
Score = 54.7 bits (130), Expect = 1e-06
Identities = 24/68 (35%), Positives = 41/68 (60%)
Frame = -2
Query: 692 YQPTIASPYGVDVTVTPSKLAFDGKQQSLGYEITIAVSGNPVIVDSSYSFGSITWSDGAH 513
Y+ T+ P G VTVTP L F+ + + +++ ++ + ++ Y FGS+TWSD H
Sbjct: 702 YRVTVEPPLGFQVTVTPETLVFNSTTKKVYFKVKVSTTHK---TNTGYYFGSLTWSDSLH 758
Query: 512 DVTSPIAV 489
+VT P++V
Sbjct: 759 NVTIPLSV 766
>sptr|Q9T0B5|Q9T0B5 Subtilisin-like protease-like protein.
Length = 803
Score = 54.7 bits (130), Expect = 1e-06
Identities = 24/68 (35%), Positives = 41/68 (60%)
Frame = -2
Query: 692 YQPTIASPYGVDVTVTPSKLAFDGKQQSLGYEITIAVSGNPVIVDSSYSFGSITWSDGAH 513
Y+ T+ P G VTVTP L F+ + + +++ ++ + ++ Y FGS+TWSD H
Sbjct: 727 YRVTVEPPLGFQVTVTPETLVFNSTTKKVYFKVKVSTTHK---TNTGYYFGSLTWSDSLH 783
Query: 512 DVTSPIAV 489
+VT P++V
Sbjct: 784 NVTIPLSV 791
>sptr|P93205|P93205 Serine protease, SBT2.
Length = 775
Score = 53.9 bits (128), Expect = 2e-06
Identities = 27/70 (38%), Positives = 36/70 (51%)
Frame = -2
Query: 692 YQPTIASPYGVDVTVTPSKLAFDGKQQSLGYEITIAVSGNPVIVDSSYSFGSITWSDGAH 513
Y +++ G V V P +L F K Q L Y++T V + FGS+ W DG H
Sbjct: 706 YHVVVSAFKGAVVKVEPERLNFTSKNQKLSYKVTFKT----VSRQKAPEFGSLIWKDGTH 761
Query: 512 DVTSPIAVTW 483
V SPIA+TW
Sbjct: 762 KVRSPIAITW 771
>sptr|Q9SVT4|Q9SVT4 Serine protease-like protein.
Length = 772
Score = 53.5 bits (127), Expect = 2e-06
Identities = 27/69 (39%), Positives = 41/69 (59%), Gaps = 1/69 (1%)
Frame = -2
Query: 692 YQPTIASPYGVDVTVTPSKLAFDGK-QQSLGYEITIAVSGNPVIVDSSYSFGSITWSDGA 516
Y+ I SP G+ +TV P+ L F+ ++ L + + S V+S Y FGS+TW+DG
Sbjct: 701 YKAVIESPLGITLTVNPTTLVFNSAAKRVLTFSVKAKTSHK---VNSGYFFGSLTWTDGV 757
Query: 515 HDVTSPIAV 489
HDV P++V
Sbjct: 758 HDVIIPVSV 766
>sptrnew|AAQ56790|AAQ56790 At1g32960.
Length = 777
Score = 53.5 bits (127), Expect = 2e-06
Identities = 23/68 (33%), Positives = 41/68 (60%)
Frame = -2
Query: 692 YQPTIASPYGVDVTVTPSKLAFDGKQQSLGYEITIAVSGNPVIVDSSYSFGSITWSDGAH 513
Y+ ++ P GV V VTP L F+ K S+ + + ++ + +++ Y FGS+TW+D H
Sbjct: 701 YKVSVEPPLGVRVVVTPETLVFNSKTISVSFTVRVSTTHK---INTGYYFGSLTWTDSVH 757
Query: 512 DVTSPIAV 489
+V P++V
Sbjct: 758 NVVIPLSV 765
>sptr|Q9MAP5|Q9MAP5 Third of four adjacent putative subtilase family
> (Subtilase, putative).
Length = 777
Score = 53.5 bits (127), Expect = 2e-06
Identities = 23/68 (33%), Positives = 41/68 (60%)
Frame = -2
Query: 692 YQPTIASPYGVDVTVTPSKLAFDGKQQSLGYEITIAVSGNPVIVDSSYSFGSITWSDGAH 513
Y+ ++ P GV V VTP L F+ K S+ + + ++ + +++ Y FGS+TW+D H
Sbjct: 701 YKVSVEPPLGVRVVVTPETLVFNSKTISVSFTVRVSTTHK---INTGYYFGSLTWTDSVH 757
Query: 512 DVTSPIAV 489
+V P++V
Sbjct: 758 NVVIPLSV 765
>sptr|Q93YP3|Q93YP3 Subtilisin proteinase-like.
Length = 703
Score = 53.1 bits (126), Expect = 3e-06
Identities = 27/69 (39%), Positives = 40/69 (57%), Gaps = 1/69 (1%)
Frame = -2
Query: 692 YQPTIASPYGVDVTVTPSKLAF-DGKQQSLGYEITIAVSGNPVIVDSSYSFGSITWSDGA 516
Y+ I SP G+ +TV P+ L F ++ L + + S V++ Y FGS+TWSDG
Sbjct: 632 YRAVIESPLGITLTVNPTTLVFKSAAKRVLTFSVKAKTSHK---VNTGYFFGSLTWSDGV 688
Query: 515 HDVTSPIAV 489
HDV P++V
Sbjct: 689 HDVIIPVSV 697
>sptr|Q9SVT2|Q9SVT2 Subtilisin proteinase-like.
Length = 718
Score = 53.1 bits (126), Expect = 3e-06
Identities = 27/69 (39%), Positives = 40/69 (57%), Gaps = 1/69 (1%)
Frame = -2
Query: 692 YQPTIASPYGVDVTVTPSKLAF-DGKQQSLGYEITIAVSGNPVIVDSSYSFGSITWSDGA 516
Y+ I SP G+ +TV P+ L F ++ L + + S V++ Y FGS+TWSDG
Sbjct: 647 YRAVIESPLGITLTVNPTTLVFKSAAKRVLTFSVKAKTSHK---VNTGYFFGSLTWSDGV 703
Query: 515 HDVTSPIAV 489
HDV P++V
Sbjct: 704 HDVIIPVSV 712
>sptr|Q8GUK4|Q8GUK4 Subtilisin proteinase-like.
Length = 766
Score = 53.1 bits (126), Expect = 3e-06
Identities = 27/69 (39%), Positives = 40/69 (57%), Gaps = 1/69 (1%)
Frame = -2
Query: 692 YQPTIASPYGVDVTVTPSKLAF-DGKQQSLGYEITIAVSGNPVIVDSSYSFGSITWSDGA 516
Y+ I SP G+ +TV P+ L F ++ L + + S V++ Y FGS+TWSDG
Sbjct: 695 YRAVIESPLGITLTVNPTTLVFKSAAKRVLTFSVKAKTSHK---VNTGYFFGSLTWSDGV 751
Query: 515 HDVTSPIAV 489
HDV P++V
Sbjct: 752 HDVIIPVSV 760
>sptr|Q9MAP6|Q9MAP6 Second of four adjacent putative subtilase
family>.
Length = 763
Score = 53.1 bits (126), Expect = 3e-06
Identities = 21/68 (30%), Positives = 41/68 (60%)
Frame = -2
Query: 692 YQPTIASPYGVDVTVTPSKLAFDGKQQSLGYEITIAVSGNPVIVDSSYSFGSITWSDGAH 513
Y+ + P G+ V VTP L F+ K +S+ + + ++ + +++ + FGS+TW+D H
Sbjct: 687 YKVLVEPPLGIQVVVTPETLVFNSKTKSVSFTVIVSTTHK---INTGFYFGSLTWTDSIH 743
Query: 512 DVTSPIAV 489
+V P++V
Sbjct: 744 NVVIPVSV 751
>sptr|Q9ZUF6|Q9ZUF6 Putative serine protease (Putative subtilisin
serine protease).
Length = 754
Score = 52.8 bits (125), Expect = 4e-06
Identities = 27/70 (38%), Positives = 40/70 (57%)
Frame = -2
Query: 692 YQPTIASPYGVDVTVTPSKLAFDGKQQSLGYEITIAVSGNPVIVDSSYSFGSITWSDGAH 513
Y+ T+ V ++V PSKL+F + Y +T VS V + + FGSITWS+ H
Sbjct: 683 YKVTVNGAPSVGISVKPSKLSFKSVGEKKRYTVTF-VSKKGVSMTNKAEFGSITWSNPQH 741
Query: 512 DVTSPIAVTW 483
+V SP+A +W
Sbjct: 742 EVRSPVAFSW 751
>sptr|Q9C7U8|Q9C7U8 Subtilisin-like protein.
Length = 753
Score = 52.8 bits (125), Expect = 4e-06
Identities = 23/68 (33%), Positives = 39/68 (57%)
Frame = -2
Query: 692 YQPTIASPYGVDVTVTPSKLAFDGKQQSLGYEITIAVSGNPVIVDSSYSFGSITWSDGAH 513
Y+P I SP G+++ V P L F + + + + S V++ + FGS+ W+DG H
Sbjct: 680 YRPVIESPLGIELDVKPKTLVFGSNITKITFSVRVKSSHR---VNTDFYFGSLCWTDGVH 736
Query: 512 DVTSPIAV 489
+VT P++V
Sbjct: 737 NVTIPVSV 744
>sptrnew|CAE01304|CAE01304 OSJNBa0020P07.21 protein.
Length = 719
Score = 52.8 bits (125), Expect = 4e-06
Identities = 27/69 (39%), Positives = 41/69 (59%), Gaps = 1/69 (1%)
Frame = -2
Query: 692 YQPTIASPYGVDVTVTPSKLAFDGKQQSLGYEITIAVSGNPVIVDSSYSFGSITWSDGA- 516
Y+ + +P GV++TV P L F +++ +++TI +G P + YSFGS+ W DG
Sbjct: 641 YKAFLYTPAGVEMTVDPPVLVFSKEKKVQSFKVTIKATGRP--IQGDYSFGSLVWHDGGI 698
Query: 515 HDVTSPIAV 489
H V PIAV
Sbjct: 699 HWVRIPIAV 707
>sptr|Q9ZSB1|Q9ZSB1 F3H7.13 protein (Putative subtilisin-like
protease).
Length = 747
Score = 52.4 bits (124), Expect = 5e-06
Identities = 24/68 (35%), Positives = 40/68 (58%)
Frame = -2
Query: 692 YQPTIASPYGVDVTVTPSKLAFDGKQQSLGYEITIAVSGNPVIVDSSYSFGSITWSDGAH 513
Y+ I P GV+V VTP++L FD + + ++ + V++ Y FGS+TW+D H
Sbjct: 671 YKVVIDPPTGVNVAVTPTELVFDSTTTKRSFTVRVSTTHK---VNTGYYFGSLTWTDTLH 727
Query: 512 DVTSPIAV 489
+V P++V
Sbjct: 728 NVAIPVSV 735
>sptrnew|CAD41662|CAD41662 OSJNBa0019K04.9 protein.
Length = 776
Score = 52.0 bits (123), Expect = 7e-06
Identities = 24/70 (34%), Positives = 35/70 (50%)
Frame = -2
Query: 692 YQPTIASPYGVDVTVTPSKLAFDGKQQSLGYEITIAVSGNPVIVDSSYSFGSITWSDGAH 513
Y + G DV V P+ L F Q L Y++T+ + FG+++WSDG H
Sbjct: 707 YHVKVTKFKGADVIVEPNTLHFVSTNQKLSYKVTVTTKA----AQKAPEFGALSWSDGVH 762
Query: 512 DVTSPIAVTW 483
V SP+ +TW
Sbjct: 763 IVRSPVVLTW 772
>sptr|Q93WQ0|Q93WQ0 Subtilisin-type protease.
Length = 766
Score = 52.0 bits (123), Expect = 7e-06
Identities = 27/69 (39%), Positives = 39/69 (56%)
Frame = -2
Query: 692 YQPTIASPYGVDVTVTPSKLAFDGKQQSLGYEITIAVSGNPVIVDSSYSFGSITWSDGAH 513
Y P + +P GV VT+TP+KL F + L Y + + + + D FGSITWS+G +
Sbjct: 700 YSPIVDAPSGVHVTLTPNKLRFTKSSKKLSYRVIFSSTLTSLKED---LFGSITWSNGKY 756
Query: 512 DVTSPIAVT 486
V SP +T
Sbjct: 757 MVRSPFVLT 765
>sptr|Q94KL9|Q94KL9 Subtilisin-like protein.
Length = 766
Score = 52.0 bits (123), Expect = 7e-06
Identities = 27/69 (39%), Positives = 39/69 (56%)
Frame = -2
Query: 692 YQPTIASPYGVDVTVTPSKLAFDGKQQSLGYEITIAVSGNPVIVDSSYSFGSITWSDGAH 513
Y P + +P GV VT+TP+KL F + L Y + + + + D FGSITWS+G +
Sbjct: 700 YSPIVDAPSGVHVTLTPNKLRFTKSSKKLSYRVIFSSTLTSLKED---LFGSITWSNGKY 756
Query: 512 DVTSPIAVT 486
V SP +T
Sbjct: 757 MVRSPFVLT 765
>sptr|Q9ZSB0|Q9ZSB0 F3H7.14 protein (Putative subtilisin-like
protease).
Length = 756
Score = 51.6 bits (122), Expect = 9e-06
Identities = 23/68 (33%), Positives = 40/68 (58%)
Frame = -2
Query: 692 YQPTIASPYGVDVTVTPSKLAFDGKQQSLGYEITIAVSGNPVIVDSSYSFGSITWSDGAH 513
Y+ I P G++V VTP++L FD + + ++ + V++ Y FGS+TW+D H
Sbjct: 680 YKVVIDPPTGINVAVTPAELVFDYTTTKRSFTVRVSTTHK---VNTGYYFGSLTWTDNMH 736
Query: 512 DVTSPIAV 489
+V P++V
Sbjct: 737 NVAIPVSV 744
>sptr|Q40764|Q40764 Antifreeze-like protein (af70).
Length = 779
Score = 51.2 bits (121), Expect = 1e-05
Identities = 24/68 (35%), Positives = 38/68 (55%)
Frame = -2
Query: 692 YQPTIASPYGVDVTVTPSKLAFDGKQQSLGYEITIAVSGNPVIVDSSYSFGSITWSDGAH 513
Y+ TI +P G++V V+P L F + L + + + + Y+FG++ WSDG H
Sbjct: 712 YKVTIDAPPGLNVKVSPEILHFSKTSKKLSFNVVFTPTN---VATKGYAFGTLVWSDGKH 768
Query: 512 DVTSPIAV 489
+V SP AV
Sbjct: 769 NVRSPFAV 776
>sptr|O65351|O65351 CUCUMISIN-like serine protease (Putative serine
protease) (Putative subtilisin serine protease ARA12).
Length = 757
Score = 50.8 bits (120), Expect = 2e-05
Identities = 25/61 (40%), Positives = 34/61 (55%)
Frame = -2
Query: 665 GVDVTVTPSKLAFDGKQQSLGYEITIAVSGNPVIVDSSYSFGSITWSDGAHDVTSPIAVT 486
GV ++V P+ L F + Y +T V + S SFGSI WSDG H V SP+A++
Sbjct: 698 GVKISVEPAVLNFKEANEKKSYTVTFTVDSSKP--SGSNSFGSIEWSDGKHVVGSPVAIS 755
Query: 485 W 483
W
Sbjct: 756 W 756
Database: /db/trembl-ebi/tmp/swall
Posted date: Sep 26, 2003 8:29 PM
Number of letters in database: 406,886,216
Number of sequences in database: 1,272,877
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 583,023,481
Number of Sequences: 1272877
Number of extensions: 13191116
Number of successful extensions: 35758
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 33663
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 35680
length of database: 406,886,216
effective HSP length: 119
effective length of database: 255,413,853
effective search space used: 28606351536
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

