BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= 3276201.2.1
(1827 letters)
Database: /db/uniprot/tmp/swall
1,395,590 sequences; 442,889,342 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
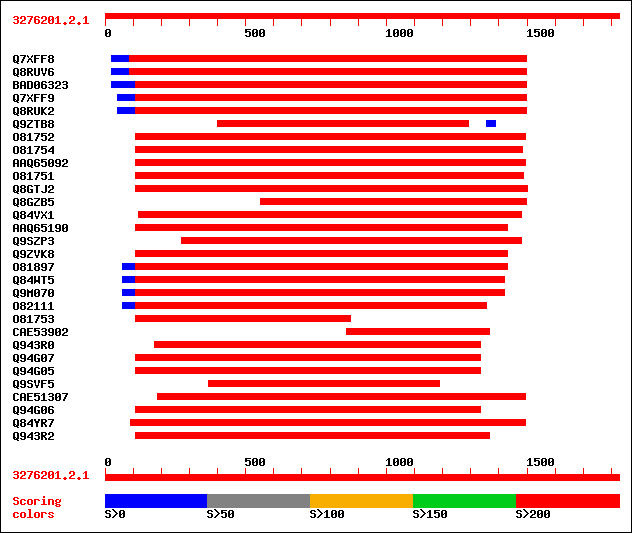
sptr|Q7XFF8|Q7XFF8 Putative 1,4-beta-xylanase. 753 0.0
sptr|Q8RUV6|Q8RUV6 Putative 1,4-beta-xylanase (Hypothetical prot... 753 0.0
sptrnew|BAD06323|BAD06323 Putative 1,4-beta-xylanase. 624 0.0
sptr|Q7XFF9|Q7XFF9 Putative 1,4-beta-xylanase. 607 e-176
sptr|Q8RUK2|Q8RUK2 Putative 1,4-beta-xylanase. 607 e-176
sptr|Q9ZTB8|Q9ZTB8 Tapetum specific protein (Tapetum-specific en... 613 e-176
sptr|O81752|O81752 Hypothetical protein. 465 e-130
sptr|O81754|O81754 Hypothetical protein. 465 e-130
sptrnew|AAQ65092|AAQ65092 At4g33840/F17I5_30. 465 e-130
sptr|O81751|O81751 Hypothetical protein. 455 e-126
sptr|Q8GTJ2|Q8GTJ2 Endoxylanase precursor (Fragment). 455 e-126
sptr|Q8GZB5|Q8GZB5 Anther endoxylanase (Fragment). 433 e-120
sptr|Q84VX1|Q84VX1 At4g38650. 371 e-101
sptrnew|AAQ65190|AAQ65190 At2g14690. 340 3e-92
sptr|Q9SZP3|Q9SZP3 Hypothetical protein. 333 4e-90
sptr|Q9ZVK8|Q9ZVK8 1,4-beta-xylan endohydrolase. 329 8e-89
sptr|O81897|O81897 Beta-xylan endohydrolase -like protein (Beta-... 310 5e-83
sptr|Q84WT5|Q84WT5 Putative glycosyl hydrolase family 10 protein. 305 1e-81
sptr|Q9M070|Q9M070 Hypothetical protein. 300 3e-80
sptr|O82111|O82111 Hypothetical protein (Fragment). 294 3e-78
sptr|O81753|O81753 Hypothetical protein. 268 2e-70
sptrnew|CAE53902|CAE53902 Putative anther endoxylanase (Fragment). 234 3e-60
sptr|Q943R0|Q943R0 Putative endo-1,4-beta-xylanase X-1. 226 9e-58
sptr|Q94G07|Q94G07 1,4-beta-D xylan xylanohydrolase (Fragment). 217 4e-55
sptr|Q94G05|Q94G05 1,4-beta-D xylan xylanohydrolase. 217 4e-55
sptr|Q9SVF5|Q9SVF5 Hypothetical protein. 216 7e-55
sptrnew|CAE51307|CAE51307 Beta-1,4-xylanase. 215 2e-54
sptr|Q94G06|Q94G06 1,4-beta-D xylan xylanohydrolase. 213 6e-54
sptr|Q84YR7|Q84YR7 Putative 1,4-beta-D xylan xylanohydrolase. 207 3e-52
sptr|Q943R2|Q943R2 Putative endo-1,4-beta-xylanase X-1. 207 4e-52
>sptr|Q7XFF8|Q7XFF8 Putative 1,4-beta-xylanase.
Length = 541
Score = 753 bits (1943), Expect = 0.0
Identities = 364/474 (76%), Positives = 410/474 (86%), Gaps = 3/474 (0%)
Frame = +2
Query: 86 GTTRYQHG--LQVSNGTSAHVRAVIKSPNGERVIAGAIVAQSGCWSMLKGGMTAYSSGPA 259
G T Y LQVS G +AHV+A +K+PNGERV+AG++ AQSGCWSMLKGGMTAYSSGP
Sbjct: 68 GDTHYSLSAWLQVSAG-AAHVKAFVKTPNGERVVAGSVSAQSGCWSMLKGGMTAYSSGPG 126
Query: 260 EIYFESHAPVDIWVDSVSLQPFTFDEWDGHTRRSADKVRRRTVKGGAMGANDKPMAHANV 439
+I+FES APVDIW+DSVSLQPFTFDEWD H ++SA KVRR TV+ GA+ PMA+A V
Sbjct: 127 QIFFESDAPVDIWMDSVSLQPFTFDEWDAHRQQSAAKVRRSTVRVVVRGADGAPMANATV 186
Query: 440 SIELLRLGFPFGNAITKEILGLPAYEKWFTSRFSVATFENEMKWYSTEWTQNHEDYRVPD 619
+ELLR GFPFGNA+TKEIL LPAYEKWFTSRF+VATFENEMKWYS EW QN+EDYRV D
Sbjct: 187 IVELLRAGFPFGNALTKEILDLPAYEKWFTSRFTVATFENEMKWYSNEWAQNNEDYRVAD 246
Query: 620 AMMNLMRKYKIKVRGHNVFWDDQNSQMQWVKPLNLAQLKAAMQKRLKSVVSPYAGKVIHW 799
AM+ L +KY IK+RGHNVFWDDQNSQM+WV PLNL QLKAAMQKRLKSVV+ YAGKVIHW
Sbjct: 247 AMLKLAQKYNIKIRGHNVFWDDQNSQMKWVTPLNLDQLKAAMQKRLKSVVTRYAGKVIHW 306
Query: 800 DVVNENLHFNFFETKLGPMASAQIYQQVGQLDRNAILFMNEFNTLEQPGDPNPVPAKYVA 979
DVVNENLHFNFFETKLGP AS IY QVG LD+NAILFMNEFNTLEQPGDPNPVP+KYVA
Sbjct: 307 DVVNENLHFNFFETKLGPNASPMIYNQVGALDKNAILFMNEFNTLEQPGDPNPVPSKYVA 366
Query: 980 KMNQIRGYAGNGGLKLGVGLESHFSTPNIPYMRSSLDTLAKLKLPMWLTEVDVVKSPNQV 1159
KM QI+ Y GN LKLGVGLESHFSTPNIPYMRS+LDTLA+LKLPMWLTEVDVVK PNQV
Sbjct: 367 KMKQIQSYPGNSALKLGVGLESHFSTPNIPYMRSALDTLAQLKLPMWLTEVDVVKGPNQV 426
Query: 1160 KYLEQVLREGFAHPNVDGIVMWAAWHARGCYVMCLTDNSFKNLPVGDLVDKLIAEWKTHR 1339
K+LEQVLREG+AHP+V+G++MWAAWHA+GCYVMCLTDNSFKNLPVG LVDKLIAEWKTH+
Sbjct: 427 KFLEQVLREGYAHPSVNGMIMWAAWHAKGCYVMCLTDNSFKNLPVGTLVDKLIAEWKTHK 486
Query: 1340 ASATTDHNGAVELHLPLGDFKFTVSHPSLKA-AAVQTMTVDNSASASQHTINVK 1498
+ATT +GAVEL LP GD+ TVSHPSL A V+ MTVD ++ AS+ +N+K
Sbjct: 487 TAATTGADGAVELDLPHGDYNLTVSHPSLGTNATVRAMTVDAASLASERLVNIK 540
Score = 45.8 bits (107), Expect = 0.002
Identities = 20/29 (68%), Positives = 23/29 (79%)
Frame = +1
Query: 22 SSSGKLLPSHSVYQRIQMQSDRHYSLSAW 108
S SG +PS SVYQ+IQ+Q D HYSLSAW
Sbjct: 49 SLSGAAVPSRSVYQKIQLQGDTHYSLSAW 77
>sptr|Q8RUV6|Q8RUV6 Putative 1,4-beta-xylanase (Hypothetical protein).
Length = 541
Score = 753 bits (1943), Expect = 0.0
Identities = 364/474 (76%), Positives = 410/474 (86%), Gaps = 3/474 (0%)
Frame = +2
Query: 86 GTTRYQHG--LQVSNGTSAHVRAVIKSPNGERVIAGAIVAQSGCWSMLKGGMTAYSSGPA 259
G T Y LQVS G +AHV+A +K+PNGERV+AG++ AQSGCWSMLKGGMTAYSSGP
Sbjct: 68 GDTHYSLSAWLQVSAG-AAHVKAFVKTPNGERVVAGSVSAQSGCWSMLKGGMTAYSSGPG 126
Query: 260 EIYFESHAPVDIWVDSVSLQPFTFDEWDGHTRRSADKVRRRTVKGGAMGANDKPMAHANV 439
+I+FES APVDIW+DSVSLQPFTFDEWD H ++SA KVRR TV+ GA+ PMA+A V
Sbjct: 127 QIFFESDAPVDIWMDSVSLQPFTFDEWDAHRQQSAAKVRRSTVRVVVRGADGAPMANATV 186
Query: 440 SIELLRLGFPFGNAITKEILGLPAYEKWFTSRFSVATFENEMKWYSTEWTQNHEDYRVPD 619
+ELLR GFPFGNA+TKEIL LPAYEKWFTSRF+VATFENEMKWYS EW QN+EDYRV D
Sbjct: 187 IVELLRAGFPFGNALTKEILDLPAYEKWFTSRFTVATFENEMKWYSNEWAQNNEDYRVAD 246
Query: 620 AMMNLMRKYKIKVRGHNVFWDDQNSQMQWVKPLNLAQLKAAMQKRLKSVVSPYAGKVIHW 799
AM+ L +KY IK+RGHNVFWDDQNSQM+WV PLNL QLKAAMQKRLKSVV+ YAGKVIHW
Sbjct: 247 AMLKLAQKYNIKIRGHNVFWDDQNSQMKWVTPLNLDQLKAAMQKRLKSVVTRYAGKVIHW 306
Query: 800 DVVNENLHFNFFETKLGPMASAQIYQQVGQLDRNAILFMNEFNTLEQPGDPNPVPAKYVA 979
DVVNENLHFNFFETKLGP AS IY QVG LD+NAILFMNEFNTLEQPGDPNPVP+KYVA
Sbjct: 307 DVVNENLHFNFFETKLGPNASPMIYNQVGALDKNAILFMNEFNTLEQPGDPNPVPSKYVA 366
Query: 980 KMNQIRGYAGNGGLKLGVGLESHFSTPNIPYMRSSLDTLAKLKLPMWLTEVDVVKSPNQV 1159
KM QI+ Y GN LKLGVGLESHFSTPNIPYMRS+LDTLA+LKLPMWLTEVDVVK PNQV
Sbjct: 367 KMKQIQSYPGNSALKLGVGLESHFSTPNIPYMRSALDTLAQLKLPMWLTEVDVVKGPNQV 426
Query: 1160 KYLEQVLREGFAHPNVDGIVMWAAWHARGCYVMCLTDNSFKNLPVGDLVDKLIAEWKTHR 1339
K+LEQVLREG+AHP+V+G++MWAAWHA+GCYVMCLTDNSFKNLPVG LVDKLIAEWKTH+
Sbjct: 427 KFLEQVLREGYAHPSVNGMIMWAAWHAKGCYVMCLTDNSFKNLPVGTLVDKLIAEWKTHK 486
Query: 1340 ASATTDHNGAVELHLPLGDFKFTVSHPSLKA-AAVQTMTVDNSASASQHTINVK 1498
+ATT +GAVEL LP GD+ TVSHPSL A V+ MTVD ++ AS+ +N+K
Sbjct: 487 TAATTGADGAVELDLPHGDYNLTVSHPSLGTNATVRAMTVDAASLASERLVNIK 540
Score = 45.8 bits (107), Expect = 0.002
Identities = 20/29 (68%), Positives = 23/29 (79%)
Frame = +1
Query: 22 SSSGKLLPSHSVYQRIQMQSDRHYSLSAW 108
S SG +PS SVYQ+IQ+Q D HYSLSAW
Sbjct: 49 SLSGAAVPSRSVYQKIQLQGDTHYSLSAW 77
>sptrnew|BAD06323|BAD06323 Putative 1,4-beta-xylanase.
Length = 574
Score = 624 bits (1608), Expect(2) = 0.0
Identities = 300/465 (64%), Positives = 370/465 (79%), Gaps = 2/465 (0%)
Frame = +2
Query: 110 LQVSNGTSAHVRAVIKSPNGERVIAGAIVAQSGCWSMLKGGMTAYSSGPAEIYFESHAPV 289
LQVS+GT+ VRAV K PNG + GA VA+SGCWSMLKGGMTA++SGP E++FE+ V
Sbjct: 110 LQVSSGTAV-VRAVFKDPNGAFIAGGATVARSGCWSMLKGGMTAFASGPGELFFEADGRV 168
Query: 290 DIWVDSVSLQPFTFDEWDGHTRRSADKVRRRTVKGGAMGANDKPMAHANVSIELLRLGFP 469
DIWVDSVSLQPF+F EW+ H R SA K RR VK A A+ P+ +ANVS++LLR GFP
Sbjct: 169 DIWVDSVSLQPFSFPEWEEHRRLSAGKARRSVVKVVARAADGVPLPNANVSVKLLRPGFP 228
Query: 470 FGNAITKEILGLPAYEKWFTSRFSVATFENEMKWYSTEWTQNHEDYRVPDAMMNLMRKYK 649
FGNA+TKEIL +PAYE+WF SRF+VA+FENEMKWYSTEW +NHEDY V DAM+ L +K+
Sbjct: 229 FGNAMTKEILDIPAYEQWFASRFTVASFENEMKWYSTEWMENHEDYTVADAMLRLAQKHG 288
Query: 650 IKVRGHNVFWDDQNSQMQWVKPLNLAQLKAAMQKRLKSVVSPYAGKVIHWDVVNENLHFN 829
I VRGHNV WD ++Q+ WVKPL+ +LKAA+QKR+ SVVS YAGKVI WDVVNENLH
Sbjct: 289 IAVRGHNVLWDTNDTQVSWVKPLDAQRLKAALQKRISSVVSRYAGKVIAWDVVNENLHGQ 348
Query: 830 FFETKLGPMASAQIYQQVGQLDRNAILFMNEFNTLEQPGDPNPVPAKYVAKMNQIRGYAG 1009
FFE++LG AS+++YQ+V ++DR A LFMNEF TLE+P D + +KYVAK+ QIR Y G
Sbjct: 349 FFESRLGRNASSEVYQRVARIDRTARLFMNEFGTLEEPLDAAAISSKYVAKLKQIRSYPG 408
Query: 1010 NGGLKLGVGLESHFSTPNIPYMRSSLDTLAKLKLPMWLTEVDV--VKSPNQVKYLEQVLR 1183
N G+KL VGLESHF TPNIPYMR++LD LA+L++P+WLTEVDV +P YLE+VLR
Sbjct: 409 NRGIKLAVGLESHFGTPNIPYMRATLDMLAQLRVPIWLTEVDVGPKGAPYVPVYLEEVLR 468
Query: 1184 EGFAHPNVDGIVMWAAWHARGCYVMCLTDNSFKNLPVGDLVDKLIAEWKTHRASATTDHN 1363
EG+ HPNV+G+VMWAAWHA+GC+VMCLTDN+F NLP GD VDKLIAEW+ H AT D N
Sbjct: 469 EGYGHPNVEGMVMWAAWHAQGCWVMCLTDNNFNNLPAGDRVDKLIAEWRAHPEGATMDAN 528
Query: 1364 GAVELHLPLGDFKFTVSHPSLKAAAVQTMTVDNSASASQHTINVK 1498
G EL L G++ FTV+HPSL + AV+T+TVD S+SA +HTI++K
Sbjct: 529 GVTELDLVHGEYNFTVTHPSLGSPAVRTLTVDASSSAVEHTIDIK 573
Score = 40.4 bits (93), Expect(2) = 0.0
Identities = 18/29 (62%), Positives = 22/29 (75%)
Frame = +1
Query: 22 SSSGKLLPSHSVYQRIQMQSDRHYSLSAW 108
S+S PS SVYQ+IQM++ HYSLSAW
Sbjct: 81 SASNNGQPSRSVYQKIQMETAHHYSLSAW 109
>sptr|Q7XFF9|Q7XFF9 Putative 1,4-beta-xylanase.
Length = 539
Score = 607 bits (1565), Expect(2) = e-176
Identities = 299/468 (63%), Positives = 375/468 (80%), Gaps = 5/468 (1%)
Frame = +2
Query: 110 LQVSNGTSAHVRAVIKSPNGERVIAGAIVAQSGCWSMLKGGMTAYSSGPAEIYFESHAPV 289
LQVS GT A+V AV+++P+G+ V AGA VA+SGCWSM+KGGMT+YSSG ++YFE+ A V
Sbjct: 72 LQVSAGT-ANVMAVVRTPDGQFVAAGATVAKSGCWSMIKGGMTSYSSGQGQLYFEADAAV 130
Query: 290 DIWVDSVSLQPFTFDEWDGHTRRSADKVRRRTVKGG--AMGANDKPMAHANVSIELLRLG 463
IWVDSVSLQPFTFDEWD H ++ + RR+ G A G + P+ +A V+ ELLR G
Sbjct: 131 AIWVDSVSLQPFTFDEWDAHRQQQSAGRARRSTLGVVVARGTDGAPVPNATVTAELLRPG 190
Query: 464 FPFGNAITKEILGLPAYEKWFTSRFSVATFENEMKWYSTEWTQNHEDYRVPDAMMNLMRK 643
FPFGNA+T+EIL PAYE+WF SRF+VATFENEMKWY+TE Q HEDYRVPDAM+ L +
Sbjct: 191 FPFGNAMTREILDNPAYEQWFASRFTVATFENEMKWYATEGRQGHEDYRVPDAMLALAER 250
Query: 644 YKIKVRGHNVFWDDQNSQMQWVKPLNLAQLKAAMQKRLKSVVSPY-AGKVIHWDVVNENL 820
+ ++VRGHNVFWDDQ++QM WV+ L +L+AAM KRL+SVVS Y G+VI WDVVNENL
Sbjct: 251 HGVRVRGHNVFWDDQSTQMAWVRSLGPDELRAAMDKRLRSVVSRYGGGRVIGWDVVNENL 310
Query: 821 HFNFFETKLGPMASAQIYQQVGQLDRNAILFMNEFNTLEQPGDPNPVPAKYVAKMNQIRG 1000
H++F++ KLGP AS IY QVG++D LFMNEFNT+EQP D + +KYVAKMNQIR
Sbjct: 311 HWSFYDGKLGPDASPAIYHQVGKIDGETPLFMNEFNTVEQPVDMAAMASKYVAKMNQIRS 370
Query: 1001 YAGNGGLKLGVGLESHF-STPNIPYMRSSLDTLAKLKLPMWLTEVDVVKSPNQVKYLEQV 1177
+ GNGGLKL VGLESHF +TPNIP+MR++LDTLA+LKLP+WLTE+DV NQ ++LE+V
Sbjct: 371 FPGNGGLKLAVGLESHFGATPNIPFMRATLDTLAQLKLPIWLTEIDVANGTNQAQHLEEV 430
Query: 1178 LREGFAHPNVDGIVMWAAWHARGCYVMCLTDNSFKNLPVGDLVDKLIAEWKTHR-ASATT 1354
LREG HPNVDG+VMWAAWHA CYVMCLTD+ FKNL VGD+VDKLIAEW+TH A ATT
Sbjct: 431 LREGHGHPNVDGMVMWAAWHATACYVMCLTDDEFKNLAVGDVVDKLIAEWRTHPVAVATT 490
Query: 1355 DHNGAVELHLPLGDFKFTVSHPSLKAAAVQTMTVDNSASASQHTINVK 1498
D +G VEL L G++ TV+HPSL ++AV+T+TVD S+S+S++ I+++
Sbjct: 491 DADGVVELDLAHGEYNVTVTHPSLVSSAVRTLTVDASSSSSENAIDIR 538
Score = 36.2 bits (82), Expect(2) = e-176
Identities = 14/22 (63%), Positives = 18/22 (81%)
Frame = +1
Query: 43 PSHSVYQRIQMQSDRHYSLSAW 108
PS +VYQ +Q+Q + HYSLSAW
Sbjct: 50 PSRTVYQTVQIQPNTHYSLSAW 71
>sptr|Q8RUK2|Q8RUK2 Putative 1,4-beta-xylanase.
Length = 539
Score = 607 bits (1565), Expect(2) = e-176
Identities = 299/468 (63%), Positives = 375/468 (80%), Gaps = 5/468 (1%)
Frame = +2
Query: 110 LQVSNGTSAHVRAVIKSPNGERVIAGAIVAQSGCWSMLKGGMTAYSSGPAEIYFESHAPV 289
LQVS GT A+V AV+++P+G+ V AGA VA+SGCWSM+KGGMT+YSSG ++YFE+ A V
Sbjct: 72 LQVSAGT-ANVMAVVRTPDGQFVAAGATVAKSGCWSMIKGGMTSYSSGQGQLYFEADAAV 130
Query: 290 DIWVDSVSLQPFTFDEWDGHTRRSADKVRRRTVKGG--AMGANDKPMAHANVSIELLRLG 463
IWVDSVSLQPFTFDEWD H ++ + RR+ G A G + P+ +A V+ ELLR G
Sbjct: 131 AIWVDSVSLQPFTFDEWDAHRQQQSAGRARRSTLGVVVARGTDGAPVPNATVTAELLRPG 190
Query: 464 FPFGNAITKEILGLPAYEKWFTSRFSVATFENEMKWYSTEWTQNHEDYRVPDAMMNLMRK 643
FPFGNA+T+EIL PAYE+WF SRF+VATFENEMKWY+TE Q HEDYRVPDAM+ L +
Sbjct: 191 FPFGNAMTREILDNPAYEQWFASRFTVATFENEMKWYATEGRQGHEDYRVPDAMLALAER 250
Query: 644 YKIKVRGHNVFWDDQNSQMQWVKPLNLAQLKAAMQKRLKSVVSPY-AGKVIHWDVVNENL 820
+ ++VRGHNVFWDDQ++QM WV+ L +L+AAM KRL+SVVS Y G+VI WDVVNENL
Sbjct: 251 HGVRVRGHNVFWDDQSTQMAWVRSLGPDELRAAMDKRLRSVVSRYGGGRVIGWDVVNENL 310
Query: 821 HFNFFETKLGPMASAQIYQQVGQLDRNAILFMNEFNTLEQPGDPNPVPAKYVAKMNQIRG 1000
H++F++ KLGP AS IY QVG++D LFMNEFNT+EQP D + +KYVAKMNQIR
Sbjct: 311 HWSFYDGKLGPDASPAIYHQVGKIDGETPLFMNEFNTVEQPVDMAAMASKYVAKMNQIRS 370
Query: 1001 YAGNGGLKLGVGLESHF-STPNIPYMRSSLDTLAKLKLPMWLTEVDVVKSPNQVKYLEQV 1177
+ GNGGLKL VGLESHF +TPNIP+MR++LDTLA+LKLP+WLTE+DV NQ ++LE+V
Sbjct: 371 FPGNGGLKLAVGLESHFGATPNIPFMRATLDTLAQLKLPIWLTEIDVANGTNQAQHLEEV 430
Query: 1178 LREGFAHPNVDGIVMWAAWHARGCYVMCLTDNSFKNLPVGDLVDKLIAEWKTHR-ASATT 1354
LREG HPNVDG+VMWAAWHA CYVMCLTD+ FKNL VGD+VDKLIAEW+TH A ATT
Sbjct: 431 LREGHGHPNVDGMVMWAAWHATACYVMCLTDDEFKNLAVGDVVDKLIAEWRTHPVAVATT 490
Query: 1355 DHNGAVELHLPLGDFKFTVSHPSLKAAAVQTMTVDNSASASQHTINVK 1498
D +G VEL L G++ TV+HPSL ++AV+T+TVD S+S+S++ I+++
Sbjct: 491 DADGVVELDLAHGEYNVTVTHPSLVSSAVRTLTVDASSSSSENAIDIR 538
Score = 36.2 bits (82), Expect(2) = e-176
Identities = 14/22 (63%), Positives = 18/22 (81%)
Frame = +1
Query: 43 PSHSVYQRIQMQSDRHYSLSAW 108
PS +VYQ +Q+Q + HYSLSAW
Sbjct: 50 PSRTVYQTVQIQPNTHYSLSAW 71
>sptr|Q9ZTB8|Q9ZTB8 Tapetum specific protein (Tapetum-specific
endoxylanase).
Length = 329
Score = 613 bits (1580), Expect(2) = e-176
Identities = 292/297 (98%), Positives = 296/297 (99%)
Frame = +2
Query: 401 MGANDKPMAHANVSIELLRLGFPFGNAITKEILGLPAYEKWFTSRFSVATFENEMKWYST 580
MGANDKPMAHANVSIELLRLGFPFGNA+TKEILGLPAYEKWFTSRFSVATFENEMKWYST
Sbjct: 1 MGANDKPMAHANVSIELLRLGFPFGNAVTKEILGLPAYEKWFTSRFSVATFENEMKWYST 60
Query: 581 EWTQNHEDYRVPDAMMNLMRKYKIKVRGHNVFWDDQNSQMQWVKPLNLAQLKAAMQKRLK 760
EWTQNHEDYRVPDAMM+LMRKYKIKVRGHNVFWDDQNSQMQWVKPLNLAQLKAAMQKRLK
Sbjct: 61 EWTQNHEDYRVPDAMMSLMRKYKIKVRGHNVFWDDQNSQMQWVKPLNLAQLKAAMQKRLK 120
Query: 761 SVVSPYAGKVIHWDVVNENLHFNFFETKLGPMASAQIYQQVGQLDRNAILFMNEFNTLEQ 940
SVVSPYAGKVIHWDVVNENLHFNFFETKLGPMASAQIYQQVGQLDRNAILFMNEFNTLEQ
Sbjct: 121 SVVSPYAGKVIHWDVVNENLHFNFFETKLGPMASAQIYQQVGQLDRNAILFMNEFNTLEQ 180
Query: 941 PGDPNPVPAKYVAKMNQIRGYAGNGGLKLGVGLESHFSTPNIPYMRSSLDTLAKLKLPMW 1120
PGDPNPVPAKYVAKMNQIRGYAGNGGLKLGVGLESHFSTPNIPYMRSSLDTLAKLKLPMW
Sbjct: 181 PGDPNPVPAKYVAKMNQIRGYAGNGGLKLGVGLESHFSTPNIPYMRSSLDTLAKLKLPMW 240
Query: 1121 LTEVDVVKSPNQVKYLEQVLREGFAHPNVDGIVMWAAWHARGCYVMCLTDNSFKNLP 1291
LTEVDVVKSPNQVKYLEQVLREGFAHPNVDGIVMWA WHA+GCYVMCLT+NSFKNLP
Sbjct: 241 LTEVDVVKSPNQVKYLEQVLREGFAHPNVDGIVMWAGWHAKGCYVMCLTNNSFKNLP 297
Score = 29.6 bits (65), Expect(2) = e-176
Identities = 11/11 (100%), Positives = 11/11 (100%)
Frame = +3
Query: 1356 ITMGRWSSTCP 1388
ITMGRWSSTCP
Sbjct: 319 ITMGRWSSTCP 329
>sptr|O81752|O81752 Hypothetical protein.
Length = 669
Score = 465 bits (1197), Expect = e-130
Identities = 243/468 (51%), Positives = 311/468 (66%), Gaps = 6/468 (1%)
Frame = +2
Query: 110 LQVSNGTSAHVRAVIKSPNGERVIAGAIVAQSGCWSMLKGGMTAYSSGPAEIYFES-HAP 286
LQVS G S V AV K NGE AG++VA+S CWSMLKGG+T SGPAE++FES +
Sbjct: 201 LQVSIGKSP-VSAVFKK-NGEYKHAGSVVAESKCWSMLKGGLTVDESGPAELFFESENTM 258
Query: 287 VDIWVDSVSLQPFTFDEWDGHTRRSADKVRRRTVKGGAMGANDKPMAHANVSIELLRLGF 466
V+IWVDSVSLQPFT +EW+ H +S KVR+ TV+ M + + +A +SIE +LG+
Sbjct: 259 VEIWVDSVSLQPFTQEEWNSHHEQSIGKVRKGTVRIRVMNNKGETIPNATISIEQKKLGY 318
Query: 467 PFGNAITKEILGLPAYEKWFTSRFSVATFENEMKWYSTEWTQNHEDYRVPDAMMNLMRKY 646
PFG A+ ILG AY+ WFT RF+V TF NEMKWYSTE + EDY DAM++ + +
Sbjct: 319 PFGCAVENNILGNQAYQNWFTQRFTVTTFGNEMKWYSTERIRGQEDYSTADAMLSFFKSH 378
Query: 647 KIKVRGHNVFWDDQNSQMQWVKPLNLAQLKAAMQKRLKSVVSPYAGKVIHWDVVNENLHF 826
I VRGHNV WDD Q WV L+ L A+++R+ SVVS Y G+++ WDVVNENLHF
Sbjct: 379 GIAVRGHNVLWDDPKYQPGWVNSLSGNDLYNAVKRRVYSVVSRYKGQLLGWDVVNENLHF 438
Query: 827 NFFETKLGPMASAQIYQQVGQLDRNAILFMNEFNTLEQPGDPNPVPAKYVAKMNQIRGYA 1006
+FFE+K GP AS Y +D +FMNE+NTLEQP D PA+Y+ K+ +++
Sbjct: 439 SFFESKFGPKASYNTYTMAHAVDPRTPMFMNEYNTLEQPKDLTSSPARYLGKLRELQSIR 498
Query: 1007 GNGGLKLGVGLESHFSTPNIPYMRSSLDTLAKLKLPMWLTEVDVVKSPN-QVKYLEQVLR 1183
G + L +GLESHFSTPNIPYMRS+LDT LP+WLTE+DV PN + Y EQVLR
Sbjct: 499 VAGKIPLAIGLESHFSTPNIPYMRSALDTFGATGLPIWLTEIDVDAPPNVRANYFEQVLR 558
Query: 1184 EGFAHPNVDGIVMWAAWHARGCYVMCLTDNSFKNLPVGDLVDKLIAEWKTHRASAT--TD 1357
EG AHP V+G+VMW + GCY MCLTD +FKNLP GD+VDKL+ EW R+ T TD
Sbjct: 559 EGHAHPKVNGMVMWTGYSPSGCYRMCLTDGNFKNLPTGDVVDKLLREWGGLRSQTTGVTD 618
Query: 1358 HNGAVELHLPLGDFKFTVSHP--SLKAAAVQTMTVDNSASASQHTINV 1495
NG E L GD+ +SHP + KA+ T+T D+ +S Q ++ V
Sbjct: 619 ANGLFEAPLFHGDYDLRISHPLTNSKASYNFTLTSDDDSSQKQPSLYV 666
Score = 129 bits (323), Expect = 2e-28
Identities = 59/108 (54%), Positives = 78/108 (72%), Gaps = 1/108 (0%)
Frame = +2
Query: 983 MNQIRGYAGNGGLKLGVGLESHFSTPNIPYMRSSLDTLAKLKLPMWLTEVDVVKSPN-QV 1159
+ +++ +G ++L +GLESHF TPNIPYMRS+LD LA L +WLTE+DV P+ Q
Sbjct: 2 LKELQSIRISGYIRLAIGLESHFKTPNIPYMRSALDILAATGLLIWLTEIDVEAPPSVQA 61
Query: 1160 KYLEQVLREGFAHPNVDGIVMWAAWHARGCYVMCLTDNSFKNLPVGDL 1303
KY EQVLR+G AHP V G+V+W + GCY MCLTD +F+NLP GD+
Sbjct: 62 KYFEQVLRDGHAHPQVKGMVVWGGYSPSGCYRMCLTDGNFRNLPTGDV 109
>sptr|O81754|O81754 Hypothetical protein.
Length = 574
Score = 465 bits (1197), Expect = e-130
Identities = 238/475 (50%), Positives = 310/475 (65%), Gaps = 17/475 (3%)
Frame = +2
Query: 110 LQVSNGTSAHVRAVIKSPNGERVIAGAIVAQSGCWSMLKGGMTAYSSGPAEIYFESH--- 280
LQVS G A V+AV K NGE +AG++VA+S CWSMLKGG+T SGPAE+YFE +
Sbjct: 93 LQVSKG-KAPVKAVFKK-NGEYKLAGSVVAESKCWSMLKGGLTVDESGPAELYFEVYFSV 150
Query: 281 -----------APVDIWVDSVSLQPFTFDEWDGHTRRSADKVRRRTVKGGAMGANDKPMA 427
V+IWVDSVSLQPFT +EW+ H +S K R+RTV+ A+ + +P+
Sbjct: 151 NCCQFLRSSEDTTVEIWVDSVSLQPFTQEEWNSHHEQSIQKERKRTVRIRAVNSKGEPIP 210
Query: 428 HANVSIELLRLGFPFGNAITKEILGLPAYEKWFTSRFSVATFENEMKWYSTEWTQNHEDY 607
A +SIE +LGFPFG + K ILG AY+ WFT RF+V TF NEMKWYSTE + EDY
Sbjct: 211 KATISIEQRKLGFPFGCEVEKNILGNKAYQNWFTQRFTVTTFANEMKWYSTEVVRGKEDY 270
Query: 608 RVPDAMMNLMRKYKIKVRGHNVFWDDQNSQMQWVKPLNLAQLKAAMQKRLKSVVSPYAGK 787
DAM+ +++ + VRGHN+ W+D Q +WV L+ L A+++R+ SVVS Y G+
Sbjct: 271 STADAMLRFFKQHGVAVRGHNILWNDPKYQPKWVNALSGNDLYNAVKRRVFSVVSRYKGQ 330
Query: 788 VIHWDVVNENLHFNFFETKLGPMASAQIYQQVGQLDRNAILFMNEFNTLEQPGDPNPVPA 967
+ WDVVNENLHF++FE K+GP AS I++ D +FMNE+NTLE+ D + A
Sbjct: 331 LAGWDVVNENLHFSYFEDKMGPKASYNIFKMAQAFDPTTTMFMNEYNTLEESSDSDSSLA 390
Query: 968 KYVAKMNQIRGYAGNGGLKLGVGLESHFSTPNIPYMRSSLDTLAKLKLPMWLTEVDVVKS 1147
+Y+ K+ +IR G + LG+GLESHF TPNIPYMRS+LDTLA LP+WLTEVDV
Sbjct: 391 RYLQKLREIRSIRVCGNISLGIGLESHFKTPNIPYMRSALDTLAATGLPIWLTEVDVEAP 450
Query: 1148 PN-QVKYLEQVLREGFAHPNVDGIVMWAAWHARGCYVMCLTDNSFKNLPVGDLVDKLIAE 1324
PN Q KY EQVLREG AHP V GIV W+ + GCY MCLTD +FKN+P GD+VDKL+ E
Sbjct: 451 PNVQAKYFEQVLREGHAHPQVKGIVTWSGYSPSGCYRMCLTDGNFKNVPTGDVVDKLLHE 510
Query: 1325 WKTHRASAT--TDHNGAVELHLPLGDFKFTVSHPSLKAAAVQTMTVDNSASASQH 1483
W R T TD +G E L GD+ ++HP + A + + + S+SQ+
Sbjct: 511 WGGFRRQTTGVTDADGYFEASLFHGDYDLKIAHPLTNSKASHSFKLTSDVSSSQN 565
>sptrnew|AAQ65092|AAQ65092 At4g33840/F17I5_30.
Length = 576
Score = 465 bits (1197), Expect = e-130
Identities = 243/468 (51%), Positives = 311/468 (66%), Gaps = 6/468 (1%)
Frame = +2
Query: 110 LQVSNGTSAHVRAVIKSPNGERVIAGAIVAQSGCWSMLKGGMTAYSSGPAEIYFES-HAP 286
LQVS G S V AV K NGE AG++VA+S CWSMLKGG+T SGPAE++FES +
Sbjct: 108 LQVSIGKSP-VSAVFKK-NGEYKHAGSVVAESKCWSMLKGGLTVDESGPAELFFESENTM 165
Query: 287 VDIWVDSVSLQPFTFDEWDGHTRRSADKVRRRTVKGGAMGANDKPMAHANVSIELLRLGF 466
V+IWVDSVSLQPFT +EW+ H +S KVR+ TV+ M + + +A +SIE +LG+
Sbjct: 166 VEIWVDSVSLQPFTQEEWNSHHEQSIGKVRKGTVRIRVMNNKGETIPNATISIEQKKLGY 225
Query: 467 PFGNAITKEILGLPAYEKWFTSRFSVATFENEMKWYSTEWTQNHEDYRVPDAMMNLMRKY 646
PFG A+ ILG AY+ WFT RF+V TF NEMKWYSTE + EDY DAM++ + +
Sbjct: 226 PFGCAVENNILGNQAYQNWFTQRFTVTTFGNEMKWYSTERIRGQEDYSTADAMLSFFKSH 285
Query: 647 KIKVRGHNVFWDDQNSQMQWVKPLNLAQLKAAMQKRLKSVVSPYAGKVIHWDVVNENLHF 826
I VRGHNV WDD Q WV L+ L A+++R+ SVVS Y G+++ WDVVNENLHF
Sbjct: 286 GIAVRGHNVLWDDPKYQPGWVNSLSGNDLYNAVKRRVYSVVSRYKGQLLGWDVVNENLHF 345
Query: 827 NFFETKLGPMASAQIYQQVGQLDRNAILFMNEFNTLEQPGDPNPVPAKYVAKMNQIRGYA 1006
+FFE+K GP AS Y +D +FMNE+NTLEQP D PA+Y+ K+ +++
Sbjct: 346 SFFESKFGPKASYNTYTMAHAVDPRTPMFMNEYNTLEQPKDLTSSPARYLGKLRELQSIR 405
Query: 1007 GNGGLKLGVGLESHFSTPNIPYMRSSLDTLAKLKLPMWLTEVDVVKSPN-QVKYLEQVLR 1183
G + L +GLESHFSTPNIPYMRS+LDT LP+WLTE+DV PN + Y EQVLR
Sbjct: 406 VAGKIPLAIGLESHFSTPNIPYMRSALDTFGATGLPIWLTEIDVDAPPNVRANYFEQVLR 465
Query: 1184 EGFAHPNVDGIVMWAAWHARGCYVMCLTDNSFKNLPVGDLVDKLIAEWKTHRASAT--TD 1357
EG AHP V+G+VMW + GCY MCLTD +FKNLP GD+VDKL+ EW R+ T TD
Sbjct: 466 EGHAHPKVNGMVMWTGYSPSGCYRMCLTDGNFKNLPTGDVVDKLLREWGGLRSQTTGVTD 525
Query: 1358 HNGAVELHLPLGDFKFTVSHP--SLKAAAVQTMTVDNSASASQHTINV 1495
NG E L GD+ +SHP + KA+ T+T D+ +S Q ++ V
Sbjct: 526 ANGLFEAPLFHGDYDLRISHPLTNSKASYNFTLTSDDDSSQKQPSLYV 573
>sptr|O81751|O81751 Hypothetical protein.
Length = 544
Score = 455 bits (1170), Expect = e-126
Identities = 240/463 (51%), Positives = 306/463 (66%), Gaps = 4/463 (0%)
Frame = +2
Query: 110 LQVSNGTSAHVRAVIKSPNGERVIAGAIVAQSGCWSMLKGGMTAYSSGPAEIYFESH-AP 286
LQVS G A V AV K NGE AG++VA+S CWSMLKGG+T SGPAE++ ES
Sbjct: 77 LQVSTG-KAPVSAVFKK-NGEYKHAGSVVAESKCWSMLKGGLTVDESGPAELFVESEDTT 134
Query: 287 VDIWVDSVSLQPFTFDEWDGHTRRSADKVRRRTVKGGAMGANDKPMAHANVSIELLRLGF 466
V+IWVDSVSLQPFT DEW+ H +S D R+ V+ + + + +A+++IE RLGF
Sbjct: 135 VEIWVDSVSLQPFTQDEWNAHQEQSIDNSRKGPVRIRVVNNKGEKIPNASITIEQKRLGF 194
Query: 467 PFGNAITKEILGLPAYEKWFTSRFSVATFENEMKWYSTEWTQNHEDYRVPDAMMNLMRKY 646
PFG+A+ + ILG AY+ WFT RF+V TFENEMKWYSTE + E+Y V DAM+ ++
Sbjct: 195 PFGSAVAQNILGNQAYQNWFTQRFTVTTFENEMKWYSTESVRGIENYTVADAMLRFFNQH 254
Query: 647 KIKVRGHNVFWDDQNSQMQWVKPLNLAQLKAAMQKRLKSVVSPYAGKVIHWDVVNENLHF 826
I VRGHNV WD Q +WV L+ L A+++R+ SVVS Y G++ WDVVNENLH
Sbjct: 255 GIAVRGHNVVWDHPKYQSKWVTSLSRNDLYNAVKRRVFSVVSRYKGQLAGWDVVNENLHH 314
Query: 827 NFFETKLGPMASAQIYQQVGQLDRNAILFMNEFNTLEQPGDPNPVPAKYVAKMNQIRGYA 1006
+FFE+K GP AS I+ +D + +FMNEF TLE P D PAKY+ K+ +++
Sbjct: 315 SFFESKFGPNASNNIFAMAHAIDPSTTMFMNEFYTLEDPTDLKASPAKYLEKLRELQSIR 374
Query: 1007 GNGGLKLGVGLESHFSTPNIPYMRSSLDTLAKLKLPMWLTEVDV-VKSPNQVKYLEQVLR 1183
G + LG+GLESHFSTPNIPYMRS+LDTL LP+WLTE+DV S +Q KY EQVLR
Sbjct: 375 VRGNIPLGIGLESHFSTPNIPYMRSALDTLGATGLPIWLTEIDVKAPSSDQAKYFEQVLR 434
Query: 1184 EGFAHPNVDGIVMWAAWHARGCYVMCLTDNSFKNLPVGDLVDKLIAEWKTHRASAT--TD 1357
EG AHP+V G+V W A +A CY MCLTD +FKNLP GD+VDKLI EW R+ T TD
Sbjct: 435 EGHAHPHVKGMVTWTA-YAPNCYHMCLTDGNFKNLPTGDVVDKLIREWGGLRSQTTEVTD 493
Query: 1358 HNGAVELHLPLGDFKFTVSHPSLKAAAVQTMTVDNSASASQHT 1486
+G E L GD+ +SHP ++ T+ S +S HT
Sbjct: 494 ADGFFEASLFHGDYDLNISHPLTNSSVSHNFTL-TSDDSSLHT 535
>sptr|Q8GTJ2|Q8GTJ2 Endoxylanase precursor (Fragment).
Length = 584
Score = 455 bits (1170), Expect = e-126
Identities = 237/470 (50%), Positives = 309/470 (65%), Gaps = 6/470 (1%)
Frame = +2
Query: 110 LQVSNGTSAHVRAVIKSPNGERVIAGAIVAQSGCWSMLKGGMTAYSSGPAEIYFES-HAP 286
++VS G + V+A+ K+ +G + AGA+VA+S CWSMLKGG+T +SGPAE+YFE+ +
Sbjct: 115 IRVSEGKTP-VKAIFKTKSGYKY-AGAVVAESNCWSMLKGGLTVDASGPAELYFETDNTS 172
Query: 287 VDIWVDSVSLQPFTFDEWDGHTRRSADKVRRRTVKGGAMGANDKPMAHANVSIELLRLGF 466
V+IW+DS+SLQPFT EW H +S K+R++ V+ A+ P+ + VSI ++GF
Sbjct: 173 VEIWIDSISLQPFTQQEWKSHQDQSIKKIRKKNVRIQAVDKLGNPLPNTTVSISPKKIGF 232
Query: 467 PFGNAITKEILGLPAYEKWFTSRFSVATFENEMKWYSTEWTQNHEDYRVPDAMMNLMRKY 646
PFG AI + I+ AY+ WF+SRF+V TFENEMKW STE +Q HEDY DAM+ +K
Sbjct: 233 PFGCAINRNIVNNNAYQSWFSSRFTVTTFENEMKWASTEPSQGHEDYSTADAMVQFAKKN 292
Query: 647 KIKVRGHNVFWDDQNSQMQWVKPLNLAQLKAAMQKRLKSVVSPYAGKVIHWDVVNENLHF 826
I +RGHNVFWDD Q WV L+ L AA KR+ SV++ Y G+VI WDVVNENLHF
Sbjct: 293 GIAIRGHNVFWDDPKYQSGWVSSLSPNDLNAAATKRINSVMNRYKGQVIGWDVVNENLHF 352
Query: 827 NFFETKLGPMASAQIYQQVGQLDRNAILFMNEFNTLEQPGDPNPVPAKYVAKMNQIRGYA 1006
+FFE+KLG ASA Y + + D + LFMNE+NT+E D PAKY+ K+ I+
Sbjct: 353 SFFESKLGANASAVFYGEAHKTDPSTTLFMNEYNTVEDSRDGQATPAKYLEKLRSIQSLP 412
Query: 1007 GNGGLKLGVGLESHFST--PNIPYMRSSLDTLAKLKLPMWLTEVDVVKSPNQVKYLEQVL 1180
GNG +G+GLESHFS+ PNIPYMRS++DTLA LP+WLTEVDV NQ + LEQ+L
Sbjct: 413 GNG--NMGIGLESHFSSSPPNIPYMRSAIDTLAATGLPVWLTEVDVQSGGNQAQSLEQIL 470
Query: 1181 REGFAHPNVDGIVMWAAWHARGCYVMCLTDNSFKNLPVGDLVDKLIAEW-KTHRASATTD 1357
RE +HP V GIV+W+AW GCY MCLTDN+F NLP GD+VDKL+ EW TD
Sbjct: 471 REAHSHPKVRGIVIWSAWSPNGCYRMCLTDNNFHNLPTGDVVDKLLREWGGGATVKGKTD 530
Query: 1358 HNGAVELHLPLGDF--KFTVSHPSLKAAAVQTMTVDNSASASQHTINVKS 1501
NG + L GD+ K V+HPS +S+S HT + S
Sbjct: 531 QNGFFQSSLFHGDYEIKVQVNHPS-----------KLPSSSSHHTFKLNS 569
>sptr|Q8GZB5|Q8GZB5 Anther endoxylanase (Fragment).
Length = 318
Score = 433 bits (1114), Expect = e-120
Identities = 204/317 (64%), Positives = 251/317 (79%), Gaps = 2/317 (0%)
Frame = +2
Query: 554 ENEMKWYSTEWTQNHEDYRVPDAMMNLMRKYKIKVRGHNVFWDDQNSQMQWVKPLNLAQL 733
ENEMKWYSTEW +N EDY VPDAM+ L +++ IKVRGHNVFWD N QM WV PL+ +L
Sbjct: 1 ENEMKWYSTEWKRNREDYSVPDAMLALAQRHGIKVRGHNVFWDTNNMQMAWVNPLSADEL 60
Query: 734 KAAMQKRLKSVVSPYAGKVIHWDVVNENLHFNFFETKLGPMASAQIYQQVGQLDRNAILF 913
KAAMQKRL S+V+ YAGKVI WDVVNENLH F+E++LGP SA++YQQV ++D NA LF
Sbjct: 61 KAAMQKRLSSLVTRYAGKVIAWDVVNENLHGQFYESRLGPNVSAELYQQVAKIDTNATLF 120
Query: 914 MNEFNTLEQPGDPNPVPAKYVAKMNQIRGYAGNGGLKLGVGLESHFSTPNIPYMRSSLDT 1093
MNE++TLE D + +KY AKM QIR Y GN G+KL VGLESHF TPNIPYMR++LD
Sbjct: 121 MNEYDTLEWALDVTAMASKYAAKMEQIRSYPGNDGIKLAVGLESHFETPNIPYMRATLDM 180
Query: 1094 LAKLKLPMWLTEVDVVKS--PNQVKYLEQVLREGFAHPNVDGIVMWAAWHARGCYVMCLT 1267
LA+LK+P+WLTEVDV P QV+YLE VLREG+ HPNV+G+V+WAAWH GC+VMCLT
Sbjct: 181 LAQLKVPIWLTEVDVSPKTRPYQVEYLEDVLREGYGHPNVEGMVLWAAWHKHGCWVMCLT 240
Query: 1268 DNSFKNLPVGDLVDKLIAEWKTHRASATTDHNGAVELHLPLGDFKFTVSHPSLKAAAVQT 1447
DNSF NLP G++VDKLI EWKTH +ATTD +G EL L G+++FTV+HPSL++ T
Sbjct: 241 DNSFTNLPTGNVVDKLIDEWKTHPVAATTDAHGVAELDLVHGEYRFTVTHPSLESPMAHT 300
Query: 1448 MTVDNSASASQHTINVK 1498
+TVD S+SA +H I++K
Sbjct: 301 LTVDASSSALEHAIDIK 317
>sptr|Q84VX1|Q84VX1 At4g38650.
Length = 562
Score = 371 bits (952), Expect = e-101
Identities = 190/461 (41%), Positives = 276/461 (59%), Gaps = 7/461 (1%)
Frame = +2
Query: 119 SNGTSAHVRAVIKSPNGERVIAGAIVAQSGCWSMLKGGMTAYSSGPAEIYF----ESHAP 286
+ SAHVRA +++ N G++ A+ GCWS LKGG S I F E
Sbjct: 101 AGAASAHVRARLRADNATLNCVGSVTAKHGCWSFLKGGFLLDSPCKQSILFFETSEDDGK 160
Query: 287 VDIWVDSVSLQPFTFDEWDGHTRRSADKVRRRTVKGGAMGANDKPMAHANVSIELLRLGF 466
+ + V S SLQPFT ++W + + R+R V N + + A V++E + F
Sbjct: 161 IQLQVTSASLQPFTQEQWRNNQDYFINTARKRAVTIHVSKENGESVEGAEVTVEQISKDF 220
Query: 467 PFGNAITKEILGLPAYEKWFTSRFSVATFENEMKWYSTEWTQNHEDYRVPDAMMNLMRKY 646
G+AI+K ILG Y++WF RF FENE+KWY+TE Q +Y + D MMN +R
Sbjct: 221 SIGSAISKTILGNIPYQEWFVKRFDATVFENELKWYATEPDQGKLNYTLADKMMNFVRAN 280
Query: 647 KIKVRGHNVFWDDQNSQMQWVKPLNLAQLKAAMQKRLKSVVSPYAGKVIHWDVVNENLHF 826
+I RGHN+FW+D WV+ L L++A+ +R+KS+++ Y G+ +HWDV NE LHF
Sbjct: 281 RIIARGHNIFWEDPKYNPDWVRNLTGEDLRSAVNRRIKSLMTRYRGEFVHWDVSNEMLHF 340
Query: 827 NFFETKLGPMASAQIYQQVGQLDRNAILFMNEFNTLEQPGDPNPVPAKYVAKMNQIRGYA 1006
+F+ET+LG AS + ++D A LF N+FN +E D +Y+A++ +++ Y
Sbjct: 341 DFYETRLGKNASYGFFAAAREIDSLATLFFNDFNVVETCSDEKSTVDEYIARVRELQRY- 399
Query: 1007 GNGGLKLGVGLESHFSTPNIPYMRSSLDTLAKLKLPMWLTEVDVVKS---PNQVKYLEQV 1177
+G G+GLE HF+TPN+ MR+ LD LA L+LP+WLTE+D+ S +Q YLEQV
Sbjct: 400 -DGVRMDGIGLEGHFTTPNVALMRAILDKLATLQLPIWLTEIDISSSLDHRSQAIYLEQV 458
Query: 1178 LREGFAHPNVDGIVMWAAWHARGCYVMCLTDNSFKNLPVGDLVDKLIAEWKTHRASATTD 1357
LREGF+HP+V+GI++W A H GCY MCLTD+ F+NLP GD+VD+ + EWKT ATTD
Sbjct: 459 LREGFSHPSVNGIMLWTALHPNGCYQMCLTDDKFRNLPAGDVVDQKLLEWKTGEVKATTD 518
Query: 1358 HNGAVELHLPLGDFKFTVSHPSLKAAAVQTMTVDNSASASQ 1480
+G+ LG+++ + + Q TV++S S SQ
Sbjct: 519 DHGSFSFFGFLGEYRVGIMY--------QGKTVNSSFSLSQ 551
>sptrnew|AAQ65190|AAQ65190 At2g14690.
Length = 529
Score = 340 bits (873), Expect = 3e-92
Identities = 179/457 (39%), Positives = 262/457 (57%), Gaps = 17/457 (3%)
Frame = +2
Query: 110 LQVSNGTSAHVRAVIKSPNGERVIAGAIVAQSGCWSMLKGGMTAYSSGPAEIYFESH--A 283
+++ N + V NG V G ++A+ GCWS+LKGG+TA SGP +I+FES A
Sbjct: 47 VKLRNESQRKVGMTFSEKNGRNVFGGEVMAKRGCWSLLKGGITADFSGPIDIFFESDGLA 106
Query: 284 PVDIWVDSVSLQPFTFDEWDGHTRRSADKVRRRTVKGGAMGANDKPMAHANVSIELLRLG 463
++I V +V +Q F +W + +K+R+ V+ N + + +SIE ++
Sbjct: 107 GLEISVQNVRMQRFHKTQWRLQQDQVIEKIRKNKVRFQMSFKNKSALEGSVISIEQIKPS 166
Query: 464 FPFGNAITKEILGLPAYEKWFTSRFSVATFENEMKWYSTEWTQNHEDYRVPDAMMNLMRK 643
F G A+ IL +Y +WF SRF + +F NEMKWY+TE + E+Y++ D+MM L +
Sbjct: 167 FLLGCAMNYRILESDSYREWFVSRFRLTSFTNEMKWYATEAVRGQENYKIADSMMQLAEE 226
Query: 644 YKIKVRGHNVFWDDQNSQMQWVKPLNLAQ-LKAAMQKRLKSVVSPYAGKVIHWDVVNENL 820
I V+GH V WDD+ Q WVK + + LK R+ SV+ Y G++I WDV+NEN+
Sbjct: 227 NAILVKGHTVLWDDKYWQPNWVKTITDPEDLKNVTLNRMNSVMKRYKGRLIGWDVMNENV 286
Query: 821 HFNFFETKLGPMASAQIYQQVGQLDRNAILFMNEFNTLEQPGDPNPVPAKYVAKMNQIRG 1000
HFN+FE LG ASA +Y +LD + LF+NEFNT+E D P V KM +I
Sbjct: 287 HFNYFENMLGGNASAIVYSLASKLDPDIPLFLNEFNTVEYDKDRVVSPVNVVKKMQEIVS 346
Query: 1001 YAGNGGLKLGVGLESHFST--PNIPYMRSSLDTLAKLKLPMWLTEVDVVKSPNQVKYLEQ 1174
+ GN +K G+G + HF+ PN+ YMR +LDTL L P+WLTEVD+ K P+QVKY+E
Sbjct: 347 FPGNNNIKGGIGAQGHFAPVQPNLAYMRYALDTLGSLSFPVWLTEVDMFKCPDQVKYMED 406
Query: 1175 VLREGFAHPNVDGIVMWAAWHARGCYVMCLTDNSFKNLPVGDLVDKLIAEWKTHRASATT 1354
+LRE ++HP V I+++ G + L D FKN GDL+DKL+ EWK
Sbjct: 407 ILREAYSHPAVKAIILYGGPEVSGFDKLTLADKDFKNTQAGDLIDKLLQEWKQEPVEIPI 466
Query: 1355 DHNG------------AVELHLPLGDFKFTVSHPSLK 1429
H+ + E+ L G ++ TV++PS+K
Sbjct: 467 QHHEHNDEEGGRIIGFSPEISLLHGHYRVTVTNPSMK 503
>sptr|Q9SZP3|Q9SZP3 Hypothetical protein.
Length = 433
Score = 333 bits (855), Expect = 4e-90
Identities = 171/414 (41%), Positives = 250/414 (60%), Gaps = 11/414 (2%)
Frame = +2
Query: 272 ESHAPVDIWVDSVSLQPFTFDEWDGHTRRSADKV--------RRRTVKGGAMGANDKPMA 427
E + + V S SLQPFT ++W + + V R+R V N + +
Sbjct: 19 EDDGKIQLQVTSASLQPFTQEQWRNNQDYFINTVIHTLISNARKRAVTIHVSKENGESVE 78
Query: 428 HANVSIELLRLGFPFGNAITKEILGLPAYEKWFTSRFSVATFENEMKWYSTEWTQNHEDY 607
A V++E + F G+AI+K ILG Y++WF RF FENE+KWY+TE Q +Y
Sbjct: 79 GAEVTVEQISKDFSIGSAISKTILGNIPYQEWFVKRFDATVFENELKWYATEPDQGKLNY 138
Query: 608 RVPDAMMNLMRKYKIKVRGHNVFWDDQNSQMQWVKPLNLAQLKAAMQKRLKSVVSPYAGK 787
+ D MMN +R +I RGHN+FW+D WV+ L L++A+ +R+KS+++ Y G+
Sbjct: 139 TLADKMMNFVRANRIIARGHNIFWEDPKYNPDWVRNLTGEDLRSAVNRRIKSLMTRYRGE 198
Query: 788 VIHWDVVNENLHFNFFETKLGPMASAQIYQQVGQLDRNAILFMNEFNTLEQPGDPNPVPA 967
+HWDV NE LHF+F+ET+LG AS + ++D A LF N+FN +E D
Sbjct: 199 FVHWDVSNEMLHFDFYETRLGKNASYGFFAAAREIDSLATLFFNDFNVVETCSDEKSTVD 258
Query: 968 KYVAKMNQIRGYAGNGGLKLGVGLESHFSTPNIPYMRSSLDTLAKLKLPMWLTEVDVVKS 1147
+Y+A++ +++ Y +G G+GLE HF+TPN+ MR+ LD LA L+LP+WLTE+D+ S
Sbjct: 259 EYIARVRELQRY--DGVRMDGIGLEGHFTTPNVALMRAILDKLATLQLPIWLTEIDISSS 316
Query: 1148 ---PNQVKYLEQVLREGFAHPNVDGIVMWAAWHARGCYVMCLTDNSFKNLPVGDLVDKLI 1318
+Q YLEQVLREGF+HP+V+GI++W A H GCY MCLTD+ F+NLP GD+VD+ +
Sbjct: 317 LDHRSQAIYLEQVLREGFSHPSVNGIMLWTALHPNGCYQMCLTDDKFRNLPAGDVVDQKL 376
Query: 1319 AEWKTHRASATTDHNGAVELHLPLGDFKFTVSHPSLKAAAVQTMTVDNSASASQ 1480
EWKT ATTD +G+ LG+++ + + Q TV++S S SQ
Sbjct: 377 LEWKTGEVKATTDDHGSFSFFGFLGEYRVGIMY--------QGKTVNSSFSLSQ 422
>sptr|Q9ZVK8|Q9ZVK8 1,4-beta-xylan endohydrolase.
Length = 552
Score = 329 bits (844), Expect = 8e-89
Identities = 178/480 (37%), Positives = 261/480 (54%), Gaps = 40/480 (8%)
Frame = +2
Query: 110 LQVSNGTSAHVRAVIKSPNGERVIAGAIVAQSGCWSMLKGGMTAYSSGPAEIYFESH--- 280
+++ N + V NG V G ++A+ GCWS+LKGG+TA SGP +I+FE
Sbjct: 47 VKLRNESQRKVGMTFSEKNGRNVFGGEVMAKRGCWSLLKGGITADFSGPIDIFFEVKELI 106
Query: 281 ----------------------APVDIWVDSVSLQPFTFDEWDGHTRRSADKVRRRTVKG 394
A ++I V +V +Q F +W + +K+R+ V+
Sbjct: 107 VCSFVEISSTNVGIYTKQSDGLAGLEISVQNVRMQRFHKTQWRLQQDQVIEKIRKNKVRF 166
Query: 395 GAMGANDKPMAHANVSIELLRLGFPFGNAITKEILGLPAYEKWFTSRFSVATFENEMKWY 574
N + + +SIE ++ F G A+ IL +Y +WF SRF + +F NEMKWY
Sbjct: 167 QMSFKNKSALEGSVISIEQIKPSFLLGCAMNYRILESDSYREWFVSRFRLTSFTNEMKWY 226
Query: 575 STEWTQNHEDYRVPDAMMNLMRKYKIKVRGHNVFWDDQNSQMQWVKPLNLAQ-LKAAMQK 751
+TE + E+Y++ D+MM L + I V+GH V WDD+ Q WVK + + LK
Sbjct: 227 ATEAVRGQENYKIADSMMQLAEENAILVKGHTVLWDDKYWQPNWVKTITDPEDLKNVTLN 286
Query: 752 RLKSVVSPYAGKVIHWDVVNENLHFNFFETKLGPMASAQIYQQVGQLDRNAILFMNEFNT 931
R+ SV+ Y G++I WDV+NEN+HFN+FE LG ASA +Y +LD + LF+NEFNT
Sbjct: 287 RMNSVMKRYKGRLIGWDVMNENVHFNYFENMLGGNASAIVYSLASKLDPDIPLFLNEFNT 346
Query: 932 LEQPGDPNPVPAKYVAKMNQIRGYAGNGGLKLGVGLESHFST--PNIPYMRSSLDTLAKL 1105
+E D P V KM +I + GN +K G+G + HF+ PN+ YMR +LDTL L
Sbjct: 347 VEYDKDRVVSPVNVVKKMQEIVSFPGNNNIKGGIGAQGHFAPVQPNLAYMRYALDTLGSL 406
Query: 1106 KLPMWLTEVDVVKSPNQVKYLEQVLREGFAHPNVDGIVMWAAWHARGCYVMCLTDNSFKN 1285
P+WLTEVD+ K P+QVKY+E +LRE ++HP V I+++ G + L D FKN
Sbjct: 407 SFPVWLTEVDMFKCPDQVKYMEDILREAYSHPAVKAIILYGGPEVSGFDKLTLADKDFKN 466
Query: 1286 LPVGDLVDKLIAEWKTHRASATTDHNG------------AVELHLPLGDFKFTVSHPSLK 1429
GDL+DKL+ EWK H+ + E+ L G ++ TV++PS+K
Sbjct: 467 TQAGDLIDKLLQEWKQEPVEIPIQHHEHNDEEGGRIIGFSPEISLLHGHYRVTVTNPSMK 526
>sptr|O81897|O81897 Beta-xylan endohydrolase -like protein (Beta-xylan
endohydrolase-like protein).
Length = 536
Score = 310 bits (793), Expect(2) = 5e-83
Identities = 169/455 (37%), Positives = 245/455 (53%), Gaps = 15/455 (3%)
Frame = +2
Query: 110 LQVSNGTSAHVRAVIKSPNGERVIAGAIVAQSGCWSMLKGGMTAYSSGPAEIYFE----- 274
+++ G + V V ++ NG V G + A+ CW++LKGG+ SG +I+FE
Sbjct: 56 VKLREGNNKKVGVVFRTENGRFVHGGEVRAKKRCWTLLKGGIVPDVSGSVDIFFEVQQLA 115
Query: 275 ---SHAPVDIWVDSVSLQPFTFDEWDGHTRRSADKVRRRTVKGGAMGANDKPMAHANVSI 445
I VSL+ F+ EW + +K+R+ V+ N + A +SI
Sbjct: 116 IYSDDKEAKISASDVSLKQFSKQEWKLKQDQLIEKIRKSKVRFEVTYQNKTAVKGAVISI 175
Query: 446 ELLRLGFPFGNAITKEILGLPAYEKWFTSRFSVATFENEMKWYSTEWTQNHEDYRVPDAM 625
E + F G A+ IL Y WF SRF + +F NEMKWY+TE + HE+Y D+M
Sbjct: 176 EQTKPSFLLGCAMNFRILQSEGYRNWFASRFKITSFTNEMKWYTTEKERGHENYTAADSM 235
Query: 626 MNLMRKYKIKVRGHNVFWDDQNSQMQWVKPL-NLAQLKAAMQKRLKSVVSPYAGKVIHWD 802
+ + I VRGH V WDD Q WV + + L R+ SV++ Y GK+ WD
Sbjct: 236 LKFAEENGILVRGHTVLWDDPLMQPTWVPKIEDPNDLMNVTLNRINSVMTRYKGKLTGWD 295
Query: 803 VVNENLHFNFFETKLGPMASAQIYQQVGQLDRNAILFMNEFNTLEQPGDPNPVPAKYVAK 982
VVNEN+H+++FE LG AS+ Y +LD + +F+NE+NT+E + P K K
Sbjct: 296 VVNENVHWDYFEKMLGANASSSFYNLAFKLDPDVTMFVNEYNTIENRVEVTATPVKVKEK 355
Query: 983 MNQIRGYAGNGGLKLGVGLESHF--STPNIPYMRSSLDTLAKLKLPMWLTEVDVVKSPNQ 1156
M +I Y GN +K +G + HF + PN+ YMRS+LDTL L LP+WLTEVD+ K PNQ
Sbjct: 356 MEEILAYPGNMNIKGAIGAQGHFRPTQPNLAYMRSALDTLGSLGLPIWLTEVDMPKCPNQ 415
Query: 1157 VKYLEQVLREGFAHPNVDGIVMWAAWHARGCYVMCLTDNSFKNLPVGDLVDKLIAEWKTH 1336
Y+E++LRE ++HP V GI+++A G + L D F N GD++DKL+ EW+
Sbjct: 416 EVYIEEILREAYSHPAVKGIIIFAGPEVSGFDKLTLADKYFNNTATGDVIDKLLKEWQQS 475
Query: 1337 ----RASATTDHNGAVELHLPLGDFKFTVSHPSLK 1429
+ T N E+ L G + VSHP +K
Sbjct: 476 SEIPKIFMTDSENDEEEVSLLHGHYNVNVSHPWMK 510
Score = 22.7 bits (47), Expect(2) = 5e-83
Identities = 9/16 (56%), Positives = 10/16 (62%)
Frame = +1
Query: 61 QRIQMQSDRHYSLSAW 108
QRIQ+ YS SAW
Sbjct: 40 QRIQLHEGNIYSFSAW 55
>sptr|Q84WT5|Q84WT5 Putative glycosyl hydrolase family 10 protein.
Length = 570
Score = 305 bits (781), Expect(2) = 1e-81
Identities = 165/448 (36%), Positives = 241/448 (53%), Gaps = 11/448 (2%)
Frame = +2
Query: 110 LQVSNGTSAHVRAVIKSPNGERVIAGAIVAQSGCWSMLKGGMTAYSSGPAEIYFES-HAP 286
+++ G V V ++ NG V G + A CW++LKGG+ SGP +I+FES +
Sbjct: 96 VKLREGNDKKVGVVFRTENGRLVHGGEVRANQECWTLLKGGIVPDFSGPVDIFFESENRG 155
Query: 287 VDIWVDSVSLQPFTFDEWDGHTRRSADKVRRRTVKGGAMGANDKPMAHANVSIELLRLGF 466
I +V L+ F+ +EW + +K+R+ V+ N + +S++ + F
Sbjct: 156 AKISAHNVLLKQFSKEEWKLKQDQLIEKIRKSKVRFEVTYENKTAVKGVVISLKQTKSSF 215
Query: 467 PFGNAITKEILGLPAYEKWFTSRFSVATFENEMKWYSTEWTQNHEDYRVPDAMMNLMRKY 646
G + IL Y KWF SRF + +F NEMKWY+TE + E+Y V D+M+
Sbjct: 216 LLGCGMNFRILQSQGYRKWFASRFKITSFTNEMKWYATEKARGQENYTVADSMLKFAEDN 275
Query: 647 KIKVRGHNVFWDDQNSQMQWVKPLNLAQLKAAMQ---KRLKSVVSPYAGKVIHWDVVNEN 817
I VRGH V WD+ Q WVK N+ M R+ SV+ Y GK+ WDVVNEN
Sbjct: 276 GILVRGHTVLWDNPKMQPSWVK--NIKDPNDVMNVTLNRINSVMKRYKGKLTGWDVVNEN 333
Query: 818 LHFNFFETKLGPMASAQIYQQVGQLDRNAILFMNEFNTLEQPGDPNPVPAKYVAKMNQIR 997
LH+++FE LG AS Y ++D + LF+NE+NT+E + P K M +I
Sbjct: 334 LHWDYFEKMLGANASTSFYNLAFKIDPDVRLFVNEYNTIENTKEFTATPIKVKKMMEEIL 393
Query: 998 GYAGNGGLKLGVGLESHF--STPNIPYMRSSLDTLAKLKLPMWLTEVDVVKSPNQVKYLE 1171
Y GN +K +G + HF + PN+ Y+RS+LDTL L LP+WLTEVD+ K PNQ +Y+E
Sbjct: 394 AYPGNKNMKGAIGAQGHFGPTQPNLAYIRSALDTLGSLGLPIWLTEVDMPKCPNQAQYVE 453
Query: 1172 QVLREGFAHPNVDGIVMWAAWHARGCYVMCLTDNSFKNLPVGDLVDKLIAEWKTHRASAT 1351
+LRE ++HP V GI+++ G + L D F N GD++DKL+ EW+ +
Sbjct: 454 DILREAYSHPAVKGIIIFGGPEVSGFDKLTLADKDFNNTQTGDVIDKLLKEWQQKSSEIQ 513
Query: 1352 TD-----HNGAVELHLPLGDFKFTVSHP 1420
T+ N E+ L G + VSHP
Sbjct: 514 TNFTADSDNEEEEVSLLHGHYNVNVSHP 541
Score = 22.7 bits (47), Expect(2) = 1e-81
Identities = 9/16 (56%), Positives = 10/16 (62%)
Frame = +1
Query: 61 QRIQMQSDRHYSLSAW 108
QRIQ+ YS SAW
Sbjct: 80 QRIQLHQGNIYSFSAW 95
>sptr|Q9M070|Q9M070 Hypothetical protein.
Length = 546
Score = 300 bits (769), Expect(2) = 3e-80
Identities = 165/464 (35%), Positives = 241/464 (51%), Gaps = 27/464 (5%)
Frame = +2
Query: 110 LQVSNGTSAHVRAVIKSPNGERVIAGAIVAQSGCWSMLKGGMTAYSSGPAEIYFESHAPV 289
+++ G V V ++ NG V G + A CW++LKGG+ SGP +I+FE H +
Sbjct: 56 VKLREGNDKKVGVVFRTENGRLVHGGEVRANQECWTLLKGGIVPDFSGPVDIFFEIHTYI 115
Query: 290 -----------------DIWVDSVSLQPFTFDEWDGHTRRSADKVRRRTVKGGAMGANDK 418
I +V L+ F+ +EW + +K+R+ V+ N
Sbjct: 116 LCVNVVLMRKQSENRGAKISAHNVLLKQFSKEEWKLKQDQLIEKIRKSKVRFEVTYENKT 175
Query: 419 PMAHANVSIELLRLGFPFGNAITKEILGLPAYEKWFTSRFSVATFENEMKWYSTEWTQNH 598
+ +S++ + F G + IL Y KWF SRF + +F NEMKWY+TE +
Sbjct: 176 AVKGVVISLKQTKSSFLLGCGMNFRILQSQGYRKWFASRFKITSFTNEMKWYATEKARGQ 235
Query: 599 EDYRVPDAMMNLMRKYKIKVRGHNVFWDDQNSQMQWVKPLNLAQLKAAMQ---KRLKSVV 769
E+Y V D+M+ I VRGH V WD+ Q WVK N+ M R+ SV+
Sbjct: 236 ENYTVADSMLKFAEDNGILVRGHTVLWDNPKMQPSWVK--NIKDPNDVMNVTLNRINSVM 293
Query: 770 SPYAGKVIHWDVVNENLHFNFFETKLGPMASAQIYQQVGQLDRNAILFMNEFNTLEQPGD 949
Y GK+ WDVVNENLH+++FE LG AS Y ++D + LF+NE+NT+E +
Sbjct: 294 KRYKGKLTGWDVVNENLHWDYFEKMLGANASTSFYNLAFKIDPDVRLFVNEYNTIENTKE 353
Query: 950 PNPVPAKYVAKMNQIRGYAGNGGLKLGVGLESHF--STPNIPYMRSSLDTLAKLKLPMWL 1123
P K M +I Y GN +K +G + HF + PN+ Y+RS+LDTL L LP+WL
Sbjct: 354 FTATPIKVKKMMEEILAYPGNKNMKGAIGAQGHFGPTQPNLAYIRSALDTLGSLGLPIWL 413
Query: 1124 TEVDVVKSPNQVKYLEQVLREGFAHPNVDGIVMWAAWHARGCYVMCLTDNSFKNLPVGDL 1303
TEVD+ K PNQ +Y+E +LRE ++HP V GI+++ G + L D F N GD+
Sbjct: 414 TEVDMPKCPNQAQYVEDILREAYSHPAVKGIIIFGGPEVSGFDKLTLADKDFNNTQTGDV 473
Query: 1304 VDKLIAEWKTHRASATTD-----HNGAVELHLPLGDFKFTVSHP 1420
+DKL+ EW+ + T+ N E+ L G + VSHP
Sbjct: 474 IDKLLKEWQQKSSEIQTNFTADSDNEEEEVSLLHGHYNVNVSHP 517
Score = 22.7 bits (47), Expect(2) = 3e-80
Identities = 9/16 (56%), Positives = 10/16 (62%)
Frame = +1
Query: 61 QRIQMQSDRHYSLSAW 108
QRIQ+ YS SAW
Sbjct: 40 QRIQLHQGNIYSFSAW 55
>sptr|O82111|O82111 Hypothetical protein (Fragment).
Length = 513
Score = 294 bits (752), Expect(2) = 3e-78
Identities = 157/438 (35%), Positives = 231/438 (52%), Gaps = 22/438 (5%)
Frame = +2
Query: 110 LQVSNGTSAHVRAVIKSPNGERVIAGAIVAQSGCWSMLKGGMTAYSSGPAEIYFESHAPV 289
+++ G V V ++ NG V G + A CW++LKGG+ SGP +I+FE H +
Sbjct: 56 VKLREGNDKKVGVVFRTENGRLVHGGEVRANQECWTLLKGGIVPDFSGPVDIFFEIHTYI 115
Query: 290 -----------------DIWVDSVSLQPFTFDEWDGHTRRSADKVRRRTVKGGAMGANDK 418
I +V L+ F+ +EW + +K+R+ V+ N
Sbjct: 116 LCVNVVLMRKQSENRGAKISAHNVLLKQFSKEEWKLKQDQLIEKIRKSKVRFEVTYENKT 175
Query: 419 PMAHANVSIELLRLGFPFGNAITKEILGLPAYEKWFTSRFSVATFENEMKWYSTEWTQNH 598
+ +S++ + F G + IL Y KWF SRF + +F NEMKWY+TE +
Sbjct: 176 AVKGVVISLKQTKSSFLLGCGMNFRILQSQGYRKWFASRFKITSFTNEMKWYATEKARGQ 235
Query: 599 EDYRVPDAMMNLMRKYKIKVRGHNVFWDDQNSQMQWVKPLNLAQLKAAMQ---KRLKSVV 769
E+Y V D+M+ I VRGH V WD+ Q WVK N+ M R+ SV+
Sbjct: 236 ENYTVADSMLKFAEDNGILVRGHTVLWDNPKMQPSWVK--NIKDPNDVMNVTLNRINSVM 293
Query: 770 SPYAGKVIHWDVVNENLHFNFFETKLGPMASAQIYQQVGQLDRNAILFMNEFNTLEQPGD 949
Y GK+ WDVVNENLH+++FE LG AS Y ++D + LF+NE+NT+E +
Sbjct: 294 KRYKGKLTGWDVVNENLHWDYFEKMLGANASTSFYNLAFKIDPDVRLFVNEYNTIENTKE 353
Query: 950 PNPVPAKYVAKMNQIRGYAGNGGLKLGVGLESHF--STPNIPYMRSSLDTLAKLKLPMWL 1123
P K M +I Y GN +K +G + HF + PN+ Y+RS+LDTL L LP+WL
Sbjct: 354 FTATPIKVKKMMEEILAYPGNKNMKGAIGAQGHFGPTQPNLAYIRSALDTLGSLGLPIWL 413
Query: 1124 TEVDVVKSPNQVKYLEQVLREGFAHPNVDGIVMWAAWHARGCYVMCLTDNSFKNLPVGDL 1303
TEVD+ K PNQ +Y+E +LRE ++HP V GI+++ G + L D F N GD+
Sbjct: 414 TEVDMPKCPNQAQYVEDILREAYSHPAVKGIIIFGGPEVSGFDKLTLADKDFNNTQTGDV 473
Query: 1304 VDKLIAEWKTHRASATTD 1357
+DKL+ EW+ + T+
Sbjct: 474 IDKLLKEWQQKSSEIQTN 491
Score = 22.7 bits (47), Expect(2) = 3e-78
Identities = 9/16 (56%), Positives = 10/16 (62%)
Frame = +1
Query: 61 QRIQMQSDRHYSLSAW 108
QRIQ+ YS SAW
Sbjct: 40 QRIQLHQGNIYSFSAW 55
>sptr|O81753|O81753 Hypothetical protein.
Length = 371
Score = 268 bits (686), Expect = 2e-70
Identities = 139/255 (54%), Positives = 174/255 (68%), Gaps = 1/255 (0%)
Frame = +2
Query: 110 LQVSNGTSAHVRAVIKSPNGERVIAGAIVAQSGCWSMLKGGMTAYSSGPAEIYFESH-AP 286
LQVS G A VRA+ K NGE AG++VA+S CWSMLKGG+T SGPAE+YFES
Sbjct: 119 LQVSEG-KAPVRAIFKK-NGEYKNAGSVVAESKCWSMLKGGLTVDESGPAELYFESDDTM 176
Query: 287 VDIWVDSVSLQPFTFDEWDGHTRRSADKVRRRTVKGGAMGANDKPMAHANVSIELLRLGF 466
V+IWVDSVSLQPFT EW+ H +S K R+ V+ A+ + +P+ +A +SI+ RLGF
Sbjct: 177 VEIWVDSVSLQPFTQKEWNFHQEQSIYKARKGAVRIRAVDSEGQPIPNATISIQQKRLGF 236
Query: 467 PFGNAITKEILGLPAYEKWFTSRFSVATFENEMKWYSTEWTQNHEDYRVPDAMMNLMRKY 646
PFG + K ILG AYE WFT RF+V TF NEMKWYSTE + EDY DAM+ +++
Sbjct: 237 PFGCEVEKNILGNQAYENWFTQRFTVTTFANEMKWYSTEVVRGKEDYSTADAMLRFFKQH 296
Query: 647 KIKVRGHNVFWDDQNSQMQWVKPLNLAQLKAAMQKRLKSVVSPYAGKVIHWDVVNENLHF 826
+ VRGHNV WDD Q WV L+ L A+++R+ SVVS Y G++ WDVVNENLHF
Sbjct: 297 GVAVRGHNVLWDDPKYQPGWVNSLSGNDLYNAVKRRVFSVVSRYKGQLAGWDVVNENLHF 356
Query: 827 NFFETKLGPMASAQI 871
+FFE K+GP AS I
Sbjct: 357 SFFENKMGPKASYDI 371
>sptrnew|CAE53902|CAE53902 Putative anther endoxylanase (Fragment).
Length = 176
Score = 234 bits (597), Expect = 3e-60
Identities = 107/171 (62%), Positives = 134/171 (78%), Gaps = 2/171 (1%)
Frame = +2
Query: 860 SAQIYQQVGQLDRNAILFMNEFNTLEQPGDPNPVPAKYVAKMNQIRGYAGNGGLKLGVGL 1039
SA++Y +V ++D NA LFMNE+ TLE D + +KY AKM QIR Y GN G++L VGL
Sbjct: 4 SAEVYGEVAKIDTNATLFMNEYGTLESALDLTAMASKYAAKMEQIRSYPGNAGIRLAVGL 63
Query: 1040 ESHFSTPNIPYMRSSLDTLAKLKLPMWLTEVDV--VKSPNQVKYLEQVLREGFAHPNVDG 1213
ESHF TPN+PYMR++LD LA+LK+P+WLTE+DV P Q +YLE VLREG+ HPNV+G
Sbjct: 64 ESHFETPNLPYMRATLDMLAQLKVPIWLTEIDVSPKTGPYQAEYLEDVLREGYGHPNVEG 123
Query: 1214 IVMWAAWHARGCYVMCLTDNSFKNLPVGDLVDKLIAEWKTHRASATTDHNG 1366
+V+WAAWH GC+VMCLTDN+F NLP G++VDKLIAEWKTH +A TD NG
Sbjct: 124 MVLWAAWHKHGCWVMCLTDNNFTNLPTGNVVDKLIAEWKTHPVAARTDANG 174
>sptr|Q943R0|Q943R0 Putative endo-1,4-beta-xylanase X-1.
Length = 557
Score = 226 bits (576), Expect = 9e-58
Identities = 143/398 (35%), Positives = 211/398 (53%), Gaps = 11/398 (2%)
Frame = +2
Query: 176 VIAGAIVAQSGCWSMLKGGMTAYSSGPAEIYFESHAPVDIWVDSVSLQPFTFDEWDGHT- 352
V GA+ AQ+G W+ +KG +S F AP + V + LQ F D
Sbjct: 122 VEGGAVCAQAGRWTEIKGAFRLKASPCGATVFVQGAPDGVDVKVMDLQIFATDRRARFRK 181
Query: 353 -RRSADKVRRRTV--KGGAMGANDKPMAHANVSIELLRLGFPFGNAITKEILGLPAYEKW 523
R+ DKVR+R V K G G+ ++ A+V + + FPFG I ++ PA+ +
Sbjct: 182 LRKKTDKVRKRDVVLKFGGAGS----ISGASVRVMQMDSSFPFGACINGGVIQNPAFVDF 237
Query: 524 FTSRFSVATFENEMKWYSTEWTQNHEDYRVPDAMMNLMRKYKIKVRGHNVFWDDQNSQMQ 703
FT F A FENE+KWY TE Q +Y DA+++ +Y VRGH +FW N Q
Sbjct: 238 FTKHFDWAVFENELKWYWTEAQQGQLNYADADALLDFCDRYGKPVRGHCIFWAVDNVVQQ 297
Query: 704 WVKPLNLAQLKAAMQKRLKSVVSPYAGKVIHWDVVNENLHFNFFETKLGPMASAQIYQQV 883
W+K L+ QL AA+Q RL +++ YAG+ H+DV NE LH +F++ +LG +A ++++
Sbjct: 298 WIKGLDHDQLTAAVQGRLTGLLTRYAGRFPHYDVNNEMLHGSFYQDRLGDDINAFMFRET 357
Query: 884 GQLDRNAILFMNEFNTLEQPGDPNPVPAKYVAKMNQIRGYAGNGGLKLGVGLESHFSTPN 1063
+LD A LF+N++N +E DPN P KY+ QI G G+GL+ H + P
Sbjct: 358 ARLDPGATLFVNDYN-VEGGNDPNATPEKYI---EQITALQQKGAAVGGIGLQGHVTNPV 413
Query: 1064 IPYMRSSLDTLAKLKLPMWLTEVDVVKSPNQVKY--LEQVLREGFAHPNVDGIVMWA--- 1228
+ +LD LA LP+WLTE+DV +S ++ LE VLRE +AHP V+G++ W
Sbjct: 414 GEVICDALDKLATTDLPVWLTELDVCESDVDLRADDLEVVLREAYAHPAVEGVMFWGFMQ 473
Query: 1229 --AWHARGCYVMCLTDNSFKNLPVGDLVDKLIAEWKTH 1336
W C V +D + + G+ L EW +H
Sbjct: 474 GHMWRQDACLVN--SDGTVND--AGERFIDLRREWTSH 507
>sptr|Q94G07|Q94G07 1,4-beta-D xylan xylanohydrolase (Fragment).
Length = 497
Score = 217 bits (553), Expect = 4e-55
Identities = 151/424 (35%), Positives = 208/424 (49%), Gaps = 16/424 (3%)
Frame = +2
Query: 110 LQVSNGTSAHVR---AVIKSPNGERVIAGAIVAQSGCWSMLKGGMTAYSSGPAEIYFESH 280
L + GTS VR V + N V GA+ A+ G WS + G + + +
Sbjct: 30 LGAARGTSHPVRIDLGVEDNGNETLVECGAVCAKEGGWSEIMGAFRLRTEPRSAAVYVHG 89
Query: 281 AP--VDIWVDSVSLQPFTFDEWDGHTRRSADKVRRRTV---KGGAMGANDKPMAHANVSI 445
AP VD+ V + + P + DK R+R V G GA A A+V +
Sbjct: 90 APAGVDVKVMDLRVYPVDHKARFRQLKDKTDKARKRDVILKLGTPAGAGAG--AAASVRV 147
Query: 446 ELLRLGFPFGNAITKEILGLPAYEKWFTSRFSVATFENEMKWYSTEWTQNHEDYRVPDAM 625
L FPFG I ++ PA+ +FT+ F A FENE+KWY TE Q +Y DA+
Sbjct: 148 VQLDNAFPFGTCINTSVIQKPAFLDFFTNHFDWAVFENELKWYHTEVQQGQLNYADADAL 207
Query: 626 MNLMRKYKIKVRGHNVFWDDQNSQMQWVKPLNLAQLKAAMQKRLKSVVSPYAGKVIHWDV 805
+ + VRGH VFW QWVK LN QL++AMQ RL+ +VS YAG+ H+DV
Sbjct: 208 LAFCDRLGKTVRGHCVFWSVDGDVQQWVKNLNKDQLRSAMQSRLEGLVSRYAGRFKHYDV 267
Query: 806 VNENLHFNFFETKLGPM-ASAQIYQQVGQLDRNAILFMNEFNTLEQPGDPNPVPAKYVAK 982
NE LH FF +LG A ++++V +LD +LF+N++N +E DPN P KY
Sbjct: 268 NNEMLHGRFFRDRLGDEDVPAYMFKEVARLDPEPVLFVNDYN-VECGNDPNATPEKYA-- 324
Query: 983 MNQIRGYAGNGGLKLGVGLESHFSTPNIPYMRSSLDTLAKLKLPMWLTEVDVVKSP--NQ 1156
Q+ G + G+GL+ H P + ++LD LAK +P+W TE+DV + +
Sbjct: 325 -EQVAWLQSCGAVVRGIGLQGHVQNPVGEVICAALDRLAKTGVPIWFTELDVPEYDVGLR 383
Query: 1157 VKYLEQVLREGFAHPNVDGIVMW-----AAWHARGCYVMCLTDNSFKNLPVGDLVDKLIA 1321
K LE VLRE +AHP V+GIV W W L D G + L
Sbjct: 384 AKDLEVVLREAYAHPAVEGIVFWGFMQGTMWRQNA----WLVDADGTVNEAGQMFLNLQK 439
Query: 1322 EWKT 1333
EWKT
Sbjct: 440 EWKT 443
>sptr|Q94G05|Q94G05 1,4-beta-D xylan xylanohydrolase.
Length = 556
Score = 217 bits (553), Expect = 4e-55
Identities = 151/424 (35%), Positives = 208/424 (49%), Gaps = 16/424 (3%)
Frame = +2
Query: 110 LQVSNGTSAHVR---AVIKSPNGERVIAGAIVAQSGCWSMLKGGMTAYSSGPAEIYFESH 280
L + GTS VR V + N V GA+ A+ G WS + G + + +
Sbjct: 89 LGAARGTSHPVRIDLGVEDNGNETLVECGAVCAKEGGWSEIMGAFRLRTEPRSAAVYVHG 148
Query: 281 AP--VDIWVDSVSLQPFTFDEWDGHTRRSADKVRRRTV---KGGAMGANDKPMAHANVSI 445
AP VD+ V + + P + DK R+R V G GA A A+V +
Sbjct: 149 APAGVDVKVMDLRVYPVDHKARFRQLKDKTDKARKRDVILKLGTPAGAGAG--AAASVRV 206
Query: 446 ELLRLGFPFGNAITKEILGLPAYEKWFTSRFSVATFENEMKWYSTEWTQNHEDYRVPDAM 625
L FPFG I ++ PA+ +FT+ F A FENE+KWY TE Q +Y DA+
Sbjct: 207 VQLDNAFPFGTCINTSVIQKPAFLDFFTNHFDWAVFENELKWYHTEVQQGQLNYADADAL 266
Query: 626 MNLMRKYKIKVRGHNVFWDDQNSQMQWVKPLNLAQLKAAMQKRLKSVVSPYAGKVIHWDV 805
+ + VRGH VFW QWVK LN QL++AMQ RL+ +VS YAG+ H+DV
Sbjct: 267 LAFCDRLGKTVRGHCVFWSVDGDVQQWVKNLNKDQLRSAMQSRLEGLVSRYAGRFKHYDV 326
Query: 806 VNENLHFNFFETKLGPM-ASAQIYQQVGQLDRNAILFMNEFNTLEQPGDPNPVPAKYVAK 982
NE LH FF +LG A ++++V +LD +LF+N++N +E DPN P KY
Sbjct: 327 NNEMLHGRFFRDRLGDEDVPAYMFKEVARLDPEPVLFVNDYN-VECGNDPNATPEKYA-- 383
Query: 983 MNQIRGYAGNGGLKLGVGLESHFSTPNIPYMRSSLDTLAKLKLPMWLTEVDVVKSP--NQ 1156
Q+ G + G+GL+ H P + ++LD LAK +P+W TE+DV + +
Sbjct: 384 -EQVAWLQSCGAVVRGIGLQGHVQNPVGEVICAALDRLAKTGVPIWFTELDVPEYDVGLR 442
Query: 1157 VKYLEQVLREGFAHPNVDGIVMW-----AAWHARGCYVMCLTDNSFKNLPVGDLVDKLIA 1321
K LE VLRE +AHP V+GIV W W L D G + L
Sbjct: 443 AKDLEVVLREAYAHPAVEGIVFWGFMQGTMWRQNA----WLVDADGTVNEAGQMFLNLQK 498
Query: 1322 EWKT 1333
EWKT
Sbjct: 499 EWKT 502
>sptr|Q9SVF5|Q9SVF5 Hypothetical protein.
Length = 277
Score = 216 bits (551), Expect = 7e-55
Identities = 108/275 (39%), Positives = 166/275 (60%), Gaps = 1/275 (0%)
Frame = +2
Query: 368 KVRRRTVKGGAMGANDKPMAHANVSIELLRLGFPFGNAITKEILGLPAYEKWFTSRFSVA 547
+ R+R V N + + A V++E + FP G+AI+K ILG Y++WF RF
Sbjct: 10 QARKRAVTIHVSKENGESVEGAEVTVEQISKDFPIGSAISKTILGNIPYQEWFVKRFDAT 69
Query: 548 TFENEMKWYSTEWTQNHEDYRVPDAMMNLMRKYKIKVRGHNVFWDDQNSQMQWVKPLNLA 727
FENE+KWY+TE Q +Y + D MMNL+R +I RGHN+FW+D WV+ L
Sbjct: 70 VFENELKWYATESDQGKLNYTLADKMMNLVRANRIIARGHNIFWEDPKYNPDWVRNLTGE 129
Query: 728 QLKAAMQKRLKSVVSPYAGKVIHWDVVNENLHFNFFETKLGPMASAQIYQQVGQLDRNAI 907
L++A+ +R+KS+++ Y G+ +HWDV NE LHF+F+E++LG +D A
Sbjct: 130 DLRSAVNRRIKSLMTRYRGEFVHWDVSNEMLHFDFYESRLGK----------NVIDSLAT 179
Query: 908 LFMNEFNTLEQPGDPNPVPAKYVAKMNQIRGYAGNGGLKL-GVGLESHFSTPNIPYMRSS 1084
LF N+FN +E D +Y+A++ +++ Y G+++ G+GLE HF+TPN+ MR+
Sbjct: 180 LFFNDFNVVETCSDEKSTVDEYIARVRELQRY---DGIRMDGIGLEGHFTTPNVALMRAI 236
Query: 1085 LDTLAKLKLPMWLTEVDVVKSPNQVKYLEQVLREG 1189
LD LA L+LP+WLTE+D+ S + + + L G
Sbjct: 237 LDKLATLQLPIWLTEIDISSSLDHRSQVHKSLFSG 271
>sptrnew|CAE51307|CAE51307 Beta-1,4-xylanase.
Length = 639
Score = 215 bits (548), Expect = 2e-54
Identities = 144/448 (32%), Positives = 228/448 (50%), Gaps = 12/448 (2%)
Frame = +2
Query: 188 AIVAQSGCWSMLKG--GMTAYSSGPAEIYFESHAP-VDIWVDSVSLQPF-TFDEWDGHTR 355
+I + W L G T + E+Y E P V +VD V++ + W
Sbjct: 129 SITVEGSEWYRLSGPYSYTGTNVTNLELYIEGPQPGVSYYVDDVTVTEVGSAATWKEEAN 188
Query: 356 RSADKVRRRTVKGGAMGANDKPMAHANVSIELLRLGFPFGNAITKEILGLPAYEKWFTSR 535
+++R+R K + +N+KP++ ++ + ++ F FG+AIT + P Y ++F +
Sbjct: 189 ARIEQIRKRDPKIRIVDSNNKPVSGVSIDVRQVKHEFGFGSAITMNGIHDPRYTEFFKNN 248
Query: 536 FSVATFENEMKWYSTEWTQNHEDYRVPDAMMNLMRKYKIKVRGHNVFWDDQNSQMQWVKP 715
+ A FENE KWYS E +Q + Y D + N + IKVRGH +FW+ + Q W+K
Sbjct: 249 YEWAVFENEAKWYSNESSQGNVSYANADYLYNWCAENGIKVRGHCIFWEPEEWQPSWLKG 308
Query: 716 LNLAQLKAAMQKRLKSVVSPYAGKVIHWDVVNENLHFNFFETKLGPMASAQIYQQVGQLD 895
L L A+ RL+SVV + GK +HWDV NE LH +FF+++LG ++++ +LD
Sbjct: 309 LTGDALMKAIDARLESVVPHFRGKFLHWDVNNEMLHGDFFKSRLGESIWPYMFKRARELD 368
Query: 896 RNAILFMNEFNTLEQ-PGDPNPVPAKYVAKMNQIRGYAGNGGLKLGVGLESHFSTPNIP- 1069
+A LF+N++N + GD A + QI NG G+G++ HF P
Sbjct: 369 PDAKLFVNDYNIITYVEGD---------AYIRQIEWLLQNGAEIDGIGVQGHFDEDVEPL 419
Query: 1070 YMRSSLDTLAKLKLPMWLTEVDVVKSPN---QVKYLEQVLREGFAHPNVDGIVMWAAW-- 1234
+++ LD LA L +P+W+TE D K+P+ + + LE + R F+HP V+GI+MW W
Sbjct: 420 VVKARLDNLATLGIPIWVTEYD-SKTPDVNKRAENLENLYRIAFSHPAVEGIIMWGFWAG 478
Query: 1235 -HARGCYVMCLTDNSFKNLPVGDLVDKLIAEWKTHRASATTDHNGAVELHLPLGDFKFTV 1411
H RG + D+ + G L+ EW T S TTD GA + G ++ TV
Sbjct: 479 NHWRG-QDAAIVDHDWTVNEAGKRYQALLKEWTT-ITSGTTDSTGAFDFRGFHGTYEITV 536
Query: 1412 SHPSLKAAAVQTMTVDNSASASQHTINV 1495
S P K V+T+ + + +T V
Sbjct: 537 SVPG-KEPFVKTIELTKGNGTAVYTFTV 563
>sptr|Q94G06|Q94G06 1,4-beta-D xylan xylanohydrolase.
Length = 556
Score = 213 bits (543), Expect = 6e-54
Identities = 150/424 (35%), Positives = 206/424 (48%), Gaps = 16/424 (3%)
Frame = +2
Query: 110 LQVSNGTSAHVR---AVIKSPNGERVIAGAIVAQSGCWSMLKGGMTAYSSGPAEIYFESH 280
L + GTS VR V + N V GA+ A+ G WS + G + + +
Sbjct: 89 LGAARGTSHPVRIDLGVEDNGNETLVECGAVCAKEGGWSEIMGAFRLRTEPRSAAVYVHG 148
Query: 281 AP--VDIWVDSVSLQPFTFDEWDGHTRRSADKVRRRTV---KGGAMGANDKPMAHANVSI 445
AP VD+ V + + P + DK R+R V G GA A A+V +
Sbjct: 149 APAGVDVKVMDLRVYPVDHKARFRQLKDKTDKARKRDVILKLGTPAGAGAG--AAASVRV 206
Query: 446 ELLRLGFPFGNAITKEILGLPAYEKWFTSRFSVATFENEMKWYSTEWTQNHEDYRVPDAM 625
L FPFG I ++ PA+ +FT+ A FENE+KWY TE Q +Y DA+
Sbjct: 207 VQLDNAFPFGTCINTSVIQKPAFLDFFTNHLDWAVFENELKWYHTEVQQGQLNYADADAL 266
Query: 626 MNLMRKYKIKVRGHNVFWDDQNSQMQWVKPLNLAQLKAAMQKRLKSVVSPYAGKVIHWDV 805
+ + VRGH VFW QWVK LN QL++AMQ RL+ +VS YAG+ H+DV
Sbjct: 267 LAFCDRLGKTVRGHCVFWSVDGDVQQWVKNLNKDQLRSAMQSRLEGLVSRYAGRFKHYDV 326
Query: 806 VNENLHFNFFETKLGPM-ASAQIYQQVGQLDRNAILFMNEFNTLEQPGDPNPVPAKYVAK 982
NE LH FF +LG A ++++V +LD LF+N++N +E DPN P KY
Sbjct: 327 NNEMLHGRFFRDRLGDEDVPAYMFKEVARLDPEPALFVNDYN-VECGNDPNATPEKYA-- 383
Query: 983 MNQIRGYAGNGGLKLGVGLESHFSTPNIPYMRSSLDTLAKLKLPMWLTEVDVVKSP--NQ 1156
Q+ G + G+GL+ H P + ++LD LAK +P+W TE+DV + +
Sbjct: 384 -EQVAWLQSCGAVVRGIGLQGHVQNPVGEVICAALDRLAKTGVPIWFTELDVPEYDVGLR 442
Query: 1157 VKYLEQVLREGFAHPNVDGIVMW-----AAWHARGCYVMCLTDNSFKNLPVGDLVDKLIA 1321
K LE VLRE +AHP V+GIV W W L D G + L
Sbjct: 443 AKDLEVVLREAYAHPAVEGIVFWGFMQGTMWRQNA----WLVDADGTVNEAGQMFLNLQK 498
Query: 1322 EWKT 1333
EWKT
Sbjct: 499 EWKT 502
>sptr|Q84YR7|Q84YR7 Putative 1,4-beta-D xylan xylanohydrolase.
Length = 1101
Score = 207 bits (528), Expect = 3e-52
Identities = 149/489 (30%), Positives = 233/489 (47%), Gaps = 21/489 (4%)
Frame = +2
Query: 92 TRYQHGLQVSNGTSAHVRA---VIKSPNGERVIAGAIVAQSGCWSMLKGG---------M 235
T Y+ V G+ H R V + + + V G + A W LKG +
Sbjct: 633 TTYRVSAWVRAGSGGHGRYHVNVCLAVDHQWVNGGQVEADGDQWYELKGAFKLEKKPSKV 692
Query: 236 TAYSSGPAEIYFESHAPVDIWVDSVSLQPFTFDEWDGHTRRSADKVRRRTVKGGAMGAND 415
TAY GP VD+ V + + + DKVR+R V G++
Sbjct: 693 TAYVQGPPP-------GVDLRVMGFQIYAVDRKARFEYLKEKTDKVRKRDVILKFQGSDA 745
Query: 416 KPMAHANVSIELLRLGFPFGNAITKEILGLPAYEKWFTSRFSVATFENEMKWYSTEWTQN 595
+ +++ I+ FPFG+ I + + +F F+ A FENE+KWY TE Q
Sbjct: 746 ANLFGSSIKIQQTENSFPFGSCIGRSNIENEDLADFFMKNFNWAVFENELKWYWTEAEQG 805
Query: 596 HEDYRVPDAMMNLMRKYKIKVRGHNVFWDDQNSQMQWVKPLNLAQLKAAMQKRLKSVVSP 775
+Y+ D ++ RK+ I+VRGH +FW+ ++S W++ L+ L AA+Q RL+S++S
Sbjct: 806 RLNYKDSDELLEFCRKHNIQVRGHCLFWEVEDSVQPWIRSLHGHHLMAAIQNRLQSLLSR 865
Query: 776 YAGKVIHWDVVNENLHFNFFETKLGPMASAQIYQQVGQLDRNAILFMNEFNTLEQPGDPN 955
Y G+ H DV NE LH +F++ +LG A ++++ +LD +A+LF+N++N +E D
Sbjct: 866 YKGQFKHHDVNNEMLHGSFYQDRLGNDIRAHMFREAHKLDPSAVLFVNDYN-VEDRCDSK 924
Query: 956 PVPAKYVAKMNQIRGYAGNGGLKLGVGLESHFSTPNIPYMRSSLDTLAKLKLPMWLTEVD 1135
P K + QI G G+GL+ H + P + SLD L+ L LP+W+TE+D
Sbjct: 925 STPEKLI---EQIVDLQERGAPVGGIGLQGHITHPVGDIICDSLDKLSILGLPIWITELD 981
Query: 1136 VVKSPNQVKY--LEQVLREGFAHPNVDGIVMWAAW-------HARGCYVMCLTDNSFKNL 1288
V ++ LE LRE FAHP+V+GI++W W HA L D
Sbjct: 982 VTAENEHIRADDLEVYLREAFAHPSVEGIILWGFWELFMFREHAH------LVDVDGTIN 1035
Query: 1289 PVGDLVDKLIAEWKTHRASATTDHNGAVELHLPLGDFKFTVSHPSLKAAAVQTMTVDNSA 1468
G L EW T + DH+G ++ G + V+ PS K ++ VD
Sbjct: 1036 EAGKRYIALKQEWLT-SITGNVDHHGELKFRGYHGSYTVEVATPSGK--VTRSFVVDKDN 1092
Query: 1469 SASQHTINV 1495
+ T+N+
Sbjct: 1093 AVQVVTLNI 1101
>sptr|Q943R2|Q943R2 Putative endo-1,4-beta-xylanase X-1.
Length = 567
Score = 207 bits (527), Expect = 4e-52
Identities = 140/431 (32%), Positives = 213/431 (49%), Gaps = 11/431 (2%)
Frame = +2
Query: 107 GLQVSNGTSAHVRAVIKSPNGERVIAGAIVAQSGCWSMLKGGMTAYSSGPAEI--YFESH 280
G ++G VR + + +G V G +VA++G W + G P F
Sbjct: 108 GAGAADGCCHAVRVEVCTDDGRPVGGGVVVAEAGKWGEIMGSFRVDDDEPPRCAKVFVHG 167
Query: 281 APVDIWVDSVSLQPFTFDEWDG--HTRRSADKVRRRTVKGGAMGANDKPMAHANVSIELL 454
P + + + LQ F ++ H R+ DKVR+R V +G A V +E
Sbjct: 168 PPAGVDLKVMDLQVFAVNKIARLRHLRKKTDKVRKRDVVL-KLGRRTGGTAIRVVQVEN- 225
Query: 455 RLGFPFGNAITKEILGLPAYEKWFTSRFSVATFENEMKWYSTEWTQNHEDYRVPDAMMNL 634
FP G I K + PA+ +FT F A ENE+KWY TE Q Y D ++
Sbjct: 226 --SFPIGACINKTAIQNPAFVDFFTKHFDWAVLENELKWYYTEAVQGQVSYSDADELIAF 283
Query: 635 MRKYKIKVRGHNVFWDDQNSQMQWVKPLNLAQLKAAMQKRLKSVVSPYAGKVIHWDVVNE 814
++ VRGH +FW +N WV+ LN QL+AA++ RL+S+V+ Y G+ H++V NE
Sbjct: 284 CDRHGKPVRGHCIFWAVENVVQPWVRALNGDQLRAAVEGRLRSLVTRYGGRFPHYEVNNE 343
Query: 815 NLHFNFFETKLGPMASAQIYQQVGQLDRNAILFMNEFNTLEQPGDPNPVPAKYVAKMNQI 994
LH FF+ +LG +A+++++ Q+D + LF+N++N +E DPN P +YV + +
Sbjct: 344 MLHGAFFQQRLGDDINARMFRETAQMDPSPALFVNDYN-VESANDPNATPERYVELVTDL 402
Query: 995 RGYAGNGGLKLGVGLESHFSTPNIPYMRSSLDTLAKLKLPMWLTEVDVVKSPNQVKY--L 1168
+ G G+G++ H + P + +LD LA LP+W+TE+DV + V+ L
Sbjct: 403 Q---KRGAAVGGIGVQGHVTHPVGDVICDALDRLAVTGLPVWITELDVSAADEAVRADDL 459
Query: 1169 EQVLREGFAHPNVDGIVMWA-----AWHARGCYVMCLTDNSFKNLPVGDLVDKLIAEWKT 1333
E VLRE FAHP V+GI++W W + L D K G L EW +
Sbjct: 460 EIVLREAFAHPAVEGIMLWGFMQGNMWRSHA----HLVDADGKLNEAGHRYVGLRQEWTS 515
Query: 1334 HRASATTDHNG 1366
H A D +G
Sbjct: 516 H-ARGQVDGSG 525
Database: /db/uniprot/tmp/swall
Posted date: Mar 5, 2004 7:45 PM
Number of letters in database: 442,889,342
Number of sequences in database: 1,395,590
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,443,862,025
Number of Sequences: 1395590
Number of extensions: 32472802
Number of successful extensions: 111592
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 98875
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 110963
length of database: 442,889,342
effective HSP length: 129
effective length of database: 262,858,232
effective search space used: 125909093128
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

