BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= 3115147.2.2
(1310 letters)
Database: /db/uniprot/tmp/swall
1,395,590 sequences; 442,889,342 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
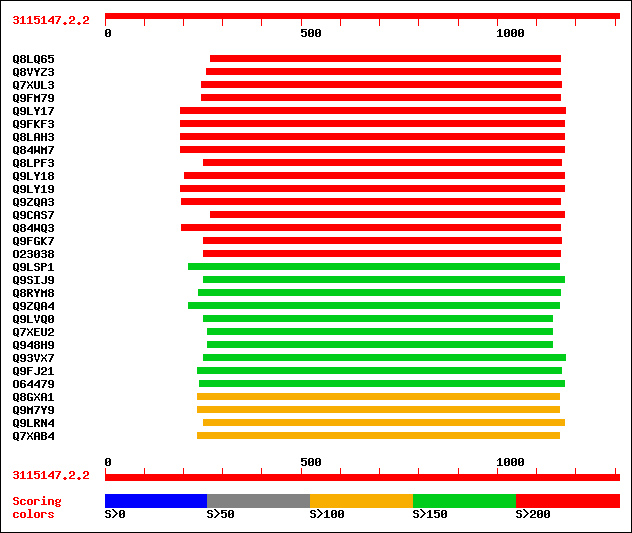
sptr|Q8LQ65|Q8LQ65 Putative pectin methylesterase. 258 1e-67
sptr|Q8VYZ3|Q8VYZ3 Putative pectin methylesterase. 251 1e-65
sptr|Q7XUL3|Q7XUL3 OSJNBa0010H02.16 protein. 247 3e-64
sptr|Q9FM79|Q9FM79 Similarity to pectin methylesterase. 243 6e-63
sptr|Q9LY17|Q9LY17 Pectin methyl-esterase-like protein. 228 2e-58
sptr|Q9FKF3|Q9FKF3 Pectin methylesterase-like protein. 228 2e-58
sptr|Q8LAH3|Q8LAH3 Pectin methyl-esterase-like protein. 224 2e-57
sptr|Q84WM7|Q84WM7 Putative pectinesterase. 223 7e-57
sptr|Q8LPF3|Q8LPF3 AT5g47500/MNJ7_9. 221 2e-56
sptr|Q9LY18|Q9LY18 Pectin methyl-esterase-like protein (Putative... 220 4e-56
sptr|Q9LY19|Q9LY19 Pectin methyl-esterase-like protein. 220 4e-56
sptr|Q9ZQA3|Q9ZQA3 Putative pectinesterase. 218 2e-55
sptr|Q9CAS7|Q9CAS7 Putative pectin methylesterase. 217 3e-55
sptr|Q84WQ3|Q84WQ3 Putative pectinesterase family protein. 215 1e-54
sptr|Q9FGK7|Q9FGK7 Pectin methylesterase-like. 214 2e-54
sptr|O23038|O23038 YUP8H12.7 protein. 202 7e-51
sptr|Q9LSP1|Q9LSP1 Pectinesterase-like protein (Pectinesterase, ... 198 2e-49
sptr|Q9SIJ9|Q9SIJ9 Putative pectinesterase. 196 5e-49
sptr|Q8RYM8|Q8RYM8 Pectin methylesterase-like protein. 194 2e-48
sptr|Q9ZQA4|Q9ZQA4 Putative pectinesterase. 194 3e-48
sptr|Q9LVQ0|Q9LVQ0 Pectin methylesterase-like protein (Putative ... 189 8e-47
sptr|Q7XEU2|Q7XEU2 Putative pectin methylesterase. 166 6e-40
sptr|Q948H9|Q948H9 Putative pectin methylesterase. 166 6e-40
sptr|Q93VX7|Q93VX7 Putative pectin methylesterase. 153 7e-36
sptr|Q9FJ21|Q9FJ21 Pectin methylesterase (EC 3.1.1.11) (AT5g4918... 153 7e-36
sptr|O64479|O64479 Putative pectinesterase. 151 2e-35
sptr|Q8GXA1|Q8GXA1 Putative pectin methylesterase. 150 6e-35
sptr|Q9M7Y9|Q9M7Y9 Pectin methylesterase, putative (EC 3.1.1.11)... 150 6e-35
sptr|Q9LRN4|Q9LRN4 Pectin methylesterase-like protein. 149 1e-34
sptr|Q7XAB4|Q7XAB4 Putative pectinesterase. 148 2e-34
>sptr|Q8LQ65|Q8LQ65 Putative pectin methylesterase.
Length = 384
Score = 258 bits (659), Expect = 1e-67
Identities = 127/299 (42%), Positives = 183/299 (61%), Gaps = 1/299 (0%)
Frame = +1
Query: 268 GDYTNITAALEDIPVSNTKRVILDLKPGAQFREKLFLNISKPFITFRSDPKKPAVVVWND 447
GD+T I AA++ +P+ N RV++ + G + EK+ ++ + FIT +V W D
Sbjct: 94 GDFTTIQAAVDSLPIINLVRVVIKVNAGT-YTEKVNISPMRAFITLEGAGADKTIVQWGD 152
Query: 448 TA-ATNGKDGKPVGTVGSATLAVESDYFTAYGVVFRNDAPLAKPGAKGGQAVAVRLFGTK 624
TA + +G+ G+P+GT SA+ AV + YF A + F+N +P+ KPGA G QAVA+R+
Sbjct: 153 TADSPSGRAGRPLGTYSSASFAVNAQYFLARNITFKNTSPVPKPGASGKQAVALRVSADN 212
Query: 625 TQIYNCTIDGGQDTLYDHKGLHYFKGCLIRGSVDFIFGFGRSFYEDCRIESVVKEVAVLT 804
C G QDTLYDH G HY+K C I GSVDFIFG S +EDC + ++ ++ LT
Sbjct: 213 AAFVGCRFLGAQDTLYDHSGRHYYKECYIEGSVDFIFGNALSLFEDCHVHAIARDYGALT 272
Query: 805 AQQRSKSIEGAIDTGFSFKNCSIGGVKGGQIYLGRAWGDSSRVVYSYTKMGEEVVPVGWD 984
AQ R +E DTGFSF NC + G G +YLGRAWG SRVV++YT M + ++P GW
Sbjct: 273 AQNRQSMLE---DTGFSFVNCRVTG--SGALYLGRAWGTFSRVVFAYTYMDDIIIPRGWY 327
Query: 985 GWQIAKPESSGIYYGEFKCFGPGADAKKKKRVGWALDLTEAQAKPFVGTHYVLGDTWIQ 1161
W E + ++YG++KC GPGA RV W+ +LT+ +AKPF+ ++ G W++
Sbjct: 328 NWGDPNRELT-VFYGQYKCTGPGASF--SGRVSWSRELTDEEAKPFISLTFIDGTEWVR 383
>sptr|Q8VYZ3|Q8VYZ3 Putative pectin methylesterase.
Length = 383
Score = 251 bits (642), Expect = 1e-65
Identities = 129/301 (42%), Positives = 179/301 (59%)
Frame = +1
Query: 259 SGKGDYTNITAALEDIPVSNTKRVILDLKPGAQFREKLFLNISKPFITFRSDPKKPAVVV 438
S KGD+T I A++ +P+ N RV++ + G ++EK+ + K FIT + + V
Sbjct: 91 SNKGDFTKIQDAIDSLPLINFVRVVIKVHAGV-YKEKVSIPPLKAFITIEGEGAEKTTVE 149
Query: 439 WNDTAATNGKDGKPVGTVGSATLAVESDYFTAYGVVFRNDAPLAKPGAKGGQAVAVRLFG 618
W DTA T G P+GT SA+ AV S +F A + FRN P+ PGA G QAVA+R+
Sbjct: 150 WGDTAQTPDSKGNPMGTYNSASFAVNSPFFVAKNITFRNTTPVPLPGAVGKQAVALRVSA 209
Query: 619 TKTQIYNCTIDGGQDTLYDHKGLHYFKGCLIRGSVDFIFGFGRSFYEDCRIESVVKEVAV 798
+ C + G QDTLYDH G HY+K C I GSVDFIFG S YE C + ++ ++
Sbjct: 210 DNAAFFGCRMLGAQDTLYDHLGRHYYKDCYIEGSVDFIFGNALSLYEGCHVHAIADKLGA 269
Query: 799 LTAQQRSKSIEGAIDTGFSFKNCSIGGVKGGQIYLGRAWGDSSRVVYSYTKMGEEVVPVG 978
+TAQ RS +E DTGFSF C + G G +YLGRAWG SRVV++YT M ++P G
Sbjct: 270 VTAQGRSSVLE---DTGFSFVKCKVTGT--GVLYLGRAWGPFSRVVFAYTYMDNIILPRG 324
Query: 979 WDGWQIAKPESSGIYYGEFKCFGPGADAKKKKRVGWALDLTEAQAKPFVGTHYVLGDTWI 1158
W W E + ++YG++KC G GA+ RV WA +LT+ +AKPF+ ++ G WI
Sbjct: 325 WYNWGDPSREMT-VFYGQYKCTGAGAN--YGGRVAWARELTDEEAKPFLSLTFIDGSEWI 381
Query: 1159 Q 1161
+
Sbjct: 382 K 382
>sptr|Q7XUL3|Q7XUL3 OSJNBa0010H02.16 protein.
Length = 344
Score = 247 bits (631), Expect = 3e-64
Identities = 128/306 (41%), Positives = 182/306 (59%)
Frame = +1
Query: 247 VVDPSGKGDYTNITAALEDIPVSNTKRVILDLKPGAQFREKLFLNISKPFITFRSDPKKP 426
VVD SG GD+ +I A+ +P +NT RVI+ + G+ + EK+ + +KP+ITF+ +
Sbjct: 45 VVDASGGGDFLSIQQAVNSVPENNTVRVIMQINAGS-YIEKVVVPATKPYITFQGAGRDV 103
Query: 427 AVVVWNDTAATNGKDGKPVGTVGSATLAVESDYFTAYGVVFRNDAPLAKPGAKGGQAVAV 606
VV W+D A+ G DG+ + T +A++ V S+YFTA + F+N AP PG +G QAVA
Sbjct: 104 TVVEWHDRASDRGPDGQQLRTYNTASVTVLSNYFTAKNISFKNTAPAPMPGMQGWQAVAF 163
Query: 607 RLFGTKTQIYNCTIDGGQDTLYDHKGLHYFKGCLIRGSVDFIFGFGRSFYEDCRIESVVK 786
R+ G K + C G QDTL D G HYF+ C I GS+DF+FG GRS Y+DC + S +
Sbjct: 164 RISGDKAFFFGCGFYGAQDTLCDDAGRHYFRDCYIEGSIDFVFGNGRSLYKDCELHSTAQ 223
Query: 787 EVAVLTAQQRSKSIEGAIDTGFSFKNCSIGGVKGGQIYLGRAWGDSSRVVYSYTKMGEEV 966
+ AQ R E TGF+F NC + G G++Y+GRA G SR+VY+YT +
Sbjct: 224 RFGSVAAQGRHDPCE---RTGFAFVNCRVTGT--GRLYVGRAMGQYSRIVYAYTYFDSVI 278
Query: 967 VPVGWDGWQIAKPESSGIYYGEFKCFGPGADAKKKKRVGWALDLTEAQAKPFVGTHYVLG 1146
P GWD W A +S ++G ++ +GPGADA V WA +L A+PF+G +V G
Sbjct: 279 APGGWDDWDHASNKSMTAFFGMYRNWGPGADA--VHGVPWARELDYFAARPFLGKSFVNG 336
Query: 1147 DTWIQP 1164
W+ P
Sbjct: 337 FHWLTP 342
>sptr|Q9FM79|Q9FM79 Similarity to pectin methylesterase.
Length = 380
Score = 243 bits (619), Expect = 6e-63
Identities = 128/307 (41%), Positives = 185/307 (60%), Gaps = 2/307 (0%)
Frame = +1
Query: 247 VVDPSGKGDYTNITAALEDIPVSNTKRVILDLKPGAQFREKLFLNISKPFITFRSDPKKP 426
VVD +G GD + A++ +P SN++RV + + PG +REK+ + SKP+I+F +
Sbjct: 83 VVDKNGGGDSVTVQGAVDMVPDSNSQRVKIFILPGI-YREKVIVPKSKPYISFIGNESYA 141
Query: 427 A--VVVWNDTAATNGKDGKPVGTVGSATLAVESDYFTAYGVVFRNDAPLAKPGAKGGQAV 600
V+ W+D A+ G DGK +GT +A++++ESD+F A + F N +A+ G +G QAV
Sbjct: 142 GDTVISWSDKASDLGCDGKELGTYRTASVSIESDFFCATAITFENTV-VAEAGEQGRQAV 200
Query: 601 AVRLFGTKTQIYNCTIDGGQDTLYDHKGLHYFKGCLIRGSVDFIFGFGRSFYEDCRIESV 780
A+R+ G K Y + G QDTL+D G HYF C I+G+VDFIFG +S Y+DC I S
Sbjct: 201 ALRIIGDKAVFYRVRVLGSQDTLFDDNGSHYFYQCYIQGNVDFIFGNAKSLYQDCDIHST 260
Query: 781 VKEVAVLTAQQRSKSIEGAIDTGFSFKNCSIGGVKGGQIYLGRAWGDSSRVVYSYTKMGE 960
K + A R E DTGFSF NC I G GQIYLGRAWG+ SR VYS + +
Sbjct: 261 AKRYGAIAAHHRDSETE---DTGFSFVNCDISGT--GQIYLGRAWGNYSRTVYSNCFIAD 315
Query: 961 EVVPVGWDGWQIAKPESSGIYYGEFKCFGPGADAKKKKRVGWALDLTEAQAKPFVGTHYV 1140
+ PVGW W+ + + + +GE+ C G GA+ + RV W+ LT + KPF+G ++
Sbjct: 316 IITPVGWSDWKHPERQRK-VMFGEYNCRGRGAE--RGGRVPWSKTLTRDEVKPFLGREFI 372
Query: 1141 LGDTWIQ 1161
GD W++
Sbjct: 373 YGDQWLR 379
>sptr|Q9LY17|Q9LY17 Pectin methyl-esterase-like protein.
Length = 361
Score = 228 bits (581), Expect = 2e-58
Identities = 127/327 (38%), Positives = 187/327 (57%)
Frame = +1
Query: 193 KEPLDKKLSEAEKKKVTYVVDPSGKGDYTNITAALEDIPVSNTKRVILDLKPGAQFREKL 372
K LD L AE + V+ G ++ + A++ IP N RVI+ L PG + EK+
Sbjct: 50 KGTLDPALEAAEAARQIITVNQKGGANFKTLNEAIKSIPTGNKNRVIIKLAPGV-YNEKV 108
Query: 373 FLNISKPFITFRSDPKKPAVVVWNDTAATNGKDGKPVGTVGSATLAVESDYFTAYGVVFR 552
++I++PFIT P V+ ++ TAA GTV SATL V ++YF A + +
Sbjct: 109 TIDIARPFITLLGQPGAETVLTYHGTAAQ-------YGTVESATLIVWAEYFQAAHLTIK 161
Query: 553 NDAPLAKPGAKGGQAVAVRLFGTKTQIYNCTIDGGQDTLYDHKGLHYFKGCLIRGSVDFI 732
N AP+ KPG++G QA+A+R+ K Y+C G QDTL D KG H+FK C I G+ DFI
Sbjct: 162 NTAPMPKPGSQG-QALAMRINADKAAFYSCRFHGFQDTLCDDKGNHFFKDCYIEGTYDFI 220
Query: 733 FGFGRSFYEDCRIESVVKEVAVLTAQQRSKSIEGAIDTGFSFKNCSIGGVKGGQIYLGRA 912
FG G S Y + ++ +V + V+TAQ R + E G++F +C + G G IYLGR+
Sbjct: 221 FGRGASLYLNTQLHAVGDGLRVITAQGRQSATE---QNGYTFVHCKVTGT-GTGIYLGRS 276
Query: 913 WGDSSRVVYSYTKMGEEVVPVGWDGWQIAKPESSGIYYGEFKCFGPGADAKKKKRVGWAL 1092
W +VVY++T+M V P GW + + ++YGE+KCFGPG+ +KRV +
Sbjct: 277 WMSHPKVVYAFTEMTSVVNPSGWRE-NLNRGYDKTVFYGEYKCFGPGSHL--EKRVPYTQ 333
Query: 1093 DLTEAQAKPFVGTHYVLGDTWIQPPPK 1173
D+ + + PF+ Y+ G TW+ PPPK
Sbjct: 334 DIDKNEVTPFLTLGYIKGSTWLLPPPK 360
>sptr|Q9FKF3|Q9FKF3 Pectin methylesterase-like protein.
Length = 364
Score = 228 bits (580), Expect = 2e-58
Identities = 128/326 (39%), Positives = 183/326 (56%)
Frame = +1
Query: 193 KEPLDKKLSEAEKKKVTYVVDPSGKGDYTNITAALEDIPVSNTKRVILDLKPGAQFREKL 372
K D L AE K V +G+G + IT A+ + NT+RVI+ + PG ++EK+
Sbjct: 52 KSTSDPALLTAEAKPRIIKVKQNGRGHFKTITEAINSVRAGNTRRVIIKIGPGV-YKEKV 110
Query: 373 FLNISKPFITFRSDPKKPAVVVWNDTAATNGKDGKPVGTVGSATLAVESDYFTAYGVVFR 552
++ SKPFIT P V+ ++ TAA GTV SATL V SDYF A ++ +
Sbjct: 111 TIDRSKPFITLYGHPNAMPVLTFDGTAAQ-------YGTVDSATLIVLSDYFMAVNIILK 163
Query: 553 NDAPLAKPGAKGGQAVAVRLFGTKTQIYNCTIDGGQDTLYDHKGLHYFKGCLIRGSVDFI 732
N AP+ KG QA+++R+ G K YNC G QDT+ D G H+FK C I G+ DFI
Sbjct: 164 NSAPMPDGKRKGAQALSMRISGNKAAFYNCKFYGYQDTICDDTGNHFFKDCYIEGTFDFI 223
Query: 733 FGFGRSFYEDCRIESVVKEVAVLTAQQRSKSIEGAIDTGFSFKNCSIGGVKGGQIYLGRA 912
FG GRS Y ++ V + V+TA + E +G+SF +C + G G IYLGR+
Sbjct: 224 FGSGRSLYLGTQLNVVGDGIRVITAHAGKSAAE---KSGYSFVHCKVTGT-GTGIYLGRS 279
Query: 913 WGDSSRVVYSYTKMGEEVVPVGWDGWQIAKPESSGIYYGEFKCFGPGADAKKKKRVGWAL 1092
W +VVY+YT M V P GW + A + + ++YGE+KC G G + K+KRV +
Sbjct: 280 WMSHPKVVYAYTDMSSVVNPSGWQENREAGRDKT-VFYGEYKCTGTG--SHKEKRVKYTQ 336
Query: 1093 DLTEAQAKPFVGTHYVLGDTWIQPPP 1170
D+ + +AK F+ Y+ G +W+ PPP
Sbjct: 337 DIDDIEAKYFISLGYIQGSSWLLPPP 362
>sptr|Q8LAH3|Q8LAH3 Pectin methyl-esterase-like protein.
Length = 342
Score = 224 bits (572), Expect = 2e-57
Identities = 125/326 (38%), Positives = 184/326 (56%)
Frame = +1
Query: 193 KEPLDKKLSEAEKKKVTYVVDPSGKGDYTNITAALEDIPVSNTKRVILDLKPGAQFREKL 372
++ LD L AE V+P G G++ +T A++ +P NTKRVI+ + PG ++REK+
Sbjct: 31 RKGLDPALVAAEAAPRIINVNPKG-GEFKTLTDAIKSVPAGNTKRVIIKMAPG-EYREKV 88
Query: 373 FLNISKPFITFRSDPKKPAVVVWNDTAATNGKDGKPVGTVGSATLAVESDYFTAYGVVFR 552
++ +KPFIT P V+ ++ TAA GTV SA+L + SDYF A +V +
Sbjct: 89 TIDRNKPFITLMGQPNAMPVITYDGTAAK-------YGTVDSASLIILSDYFMAVNIVVK 141
Query: 553 NDAPLAKPGAKGGQAVAVRLFGTKTQIYNCTIDGGQDTLYDHKGLHYFKGCLIRGSVDFI 732
N AP KG QA+++R+ G YNC G QDT+ D G H+FK C + G+ DFI
Sbjct: 142 NTAPAPDGKTKGAQALSMRISGNFAAFYNCKFYGFQDTICDDTGNHFFKDCYVEGTFDFI 201
Query: 733 FGFGRSFYEDCRIESVVKEVAVLTAQQRSKSIEGAIDTGFSFKNCSIGGVKGGQIYLGRA 912
FG G S Y ++ V + V+ A + E ++G+SF +C + G GG IYLGRA
Sbjct: 202 FGSGTSMYLGTQLHVVGDGIRVIAAHAGKSAEE---NSGYSFVHCKVTGT-GGVIYLGRA 257
Query: 913 WGDSSRVVYSYTKMGEEVVPVGWDGWQIAKPESSGIYYGEFKCFGPGADAKKKKRVGWAL 1092
W +VVY+YT+M V P GW + + + ++YGE+KC GPG + K KRV +
Sbjct: 258 WMSHPKVVYAYTEMTSVVNPTGWQENKTPAHDKT-VFYGEYKCSGPG--SHKAKRVPFTQ 314
Query: 1093 DLTEAQAKPFVGTHYVLGDTWIQPPP 1170
D+ + +A F+ Y+ G W+ PPP
Sbjct: 315 DIDDKEANRFLSLGYIQGSKWLLPPP 340
>sptr|Q84WM7|Q84WM7 Putative pectinesterase.
Length = 361
Score = 223 bits (567), Expect = 7e-57
Identities = 124/326 (38%), Positives = 183/326 (56%)
Frame = +1
Query: 193 KEPLDKKLSEAEKKKVTYVVDPSGKGDYTNITAALEDIPVSNTKRVILDLKPGAQFREKL 372
++ LD L AE V+P G G++ +T A++ +P NTKRVI+ + PG +++EK+
Sbjct: 50 RKGLDPALVAAEAAPRIINVNPKG-GEFKTLTDAIKSVPAGNTKRVIIKMAPG-EYKEKV 107
Query: 373 FLNISKPFITFRSDPKKPAVVVWNDTAATNGKDGKPVGTVGSATLAVESDYFTAYGVVFR 552
++ +KPFIT P V+ ++ TAA GTV SA+L + SDYF A +V +
Sbjct: 108 TIDRNKPFITLMGQPNAMPVITYDGTAAK-------YGTVDSASLIILSDYFMAVNIVVK 160
Query: 553 NDAPLAKPGAKGGQAVAVRLFGTKTQIYNCTIDGGQDTLYDHKGLHYFKGCLIRGSVDFI 732
N AP KG QA+++R+ G YNC G QDT+ D G H+FK C + G+ DFI
Sbjct: 161 NTAPAPDGKTKGAQALSMRISGNFAAFYNCKFYGFQDTICDDTGNHFFKDCYVEGTFDFI 220
Query: 733 FGFGRSFYEDCRIESVVKEVAVLTAQQRSKSIEGAIDTGFSFKNCSIGGVKGGQIYLGRA 912
FG G S Y ++ V + V+ A + E +G+SF +C + G GG IYLGRA
Sbjct: 221 FGSGTSMYLGTQLHVVGDGIRVIAAHAGKSAEE---KSGYSFVHCKVTGT-GGGIYLGRA 276
Query: 913 WGDSSRVVYSYTKMGEEVVPVGWDGWQIAKPESSGIYYGEFKCFGPGADAKKKKRVGWAL 1092
W +VVY+YT+M V P GW + + + ++YGE+KC GPG + K KRV +
Sbjct: 277 WMSHPKVVYAYTEMTSVVNPTGWQENKTPAHDKT-VFYGEYKCSGPG--SHKAKRVPFTQ 333
Query: 1093 DLTEAQAKPFVGTHYVLGDTWIQPPP 1170
D+ + +A F+ Y+ G W+ PPP
Sbjct: 334 DIDDKEANRFLSLGYIQGSKWLLPPP 359
>sptr|Q8LPF3|Q8LPF3 AT5g47500/MNJ7_9.
Length = 362
Score = 221 bits (564), Expect = 2e-56
Identities = 117/305 (38%), Positives = 168/305 (55%)
Frame = +1
Query: 250 VDPSGKGDYTNITAALEDIPVSNTKRVILDLKPGAQFREKLFLNISKPFITFRSDPKKPA 429
V +G + ++ A++ IP +N K + + + PG +REK+ + +KP+ITF+ +
Sbjct: 63 VSLNGHAQFRSVQDAVDSIPKNNNKSITIKIAPGF-YREKVVVPATKPYITFKGAGRDVT 121
Query: 430 VVVWNDTAATNGKDGKPVGTVGSATLAVESDYFTAYGVVFRNDAPLAKPGAKGGQAVAVR 609
+ W+D A+ G +G+ + T +A++ V ++YFTA + F N AP PG +G QAVA R
Sbjct: 122 AIEWHDRASDLGANGQQLRTYQTASVTVYANYFTARNISFTNTAPAPLPGMQGWQAVAFR 181
Query: 610 LFGTKTQIYNCTIDGGQDTLYDHKGLHYFKGCLIRGSVDFIFGFGRSFYEDCRIESVVKE 789
+ G K C G QDTL D G HYFK C I GS+DFIFG GRS Y+DC + S+
Sbjct: 182 ISGDKAFFSGCGFYGAQDTLCDDAGRHYFKECYIEGSIDFIFGNGRSMYKDCELHSIASR 241
Query: 790 VAVLTAQQRSKSIEGAIDTGFSFKNCSIGGVKGGQIYLGRAWGDSSRVVYSYTKMGEEVV 969
+ A R+ E TGF+F C + G G +Y+GRA G SR+VY+YT V
Sbjct: 242 FGSIAAHGRTCPEE---KTGFAFVGCRVTGT--GPLYVGRAMGQYSRIVYAYTYFDALVA 296
Query: 970 PVGWDGWQIAKPESSGIYYGEFKCFGPGADAKKKKRVGWALDLTEAQAKPFVGTHYVLGD 1149
GWD W +S ++G + C+GPGA A + V WA L A PF+ +V G
Sbjct: 297 HGGWDDWDHKSNKSKTAFFGVYNCYGPGAAA--TRGVSWARALDYESAHPFIAKSFVNGR 354
Query: 1150 TWIQP 1164
WI P
Sbjct: 355 HWIAP 359
>sptr|Q9LY18|Q9LY18 Pectin methyl-esterase-like protein (Putative
pectinesterase).
Length = 361
Score = 220 bits (560), Expect = 4e-56
Identities = 125/323 (38%), Positives = 180/323 (55%)
Frame = +1
Query: 202 LDKKLSEAEKKKVTYVVDPSGKGDYTNITAALEDIPVSNTKRVILDLKPGAQFREKLFLN 381
LD +L AE + +V+ +G GD+ I AA++ IP++N RVI+ L PG + EK+ ++
Sbjct: 53 LDPELEAAEASRRVIIVNQNGGGDFKTINAAIKSIPLANKNRVIIKLAPGI-YHEKVTVD 111
Query: 382 ISKPFITFRSDPKKPAVVVWNDTAATNGKDGKPVGTVGSATLAVESDYFTAYGVVFRNDA 561
+ +P++T P + + TAA GTV SATL V + F A + N +
Sbjct: 112 VGRPYVTLLGKPGAETNLTYAGTAAK-------YGTVESATLIVWATNFLAANLNIINTS 164
Query: 562 PLAKPGAKGGQAVAVRLFGTKTQIYNCTIDGGQDTLYDHKGLHYFKGCLIRGSVDFIFGF 741
P+ KPG +G QA+A+R+ G K YNC G QDTL D +G H+FK C I G+ DFIFG
Sbjct: 165 PMPKPGTQG-QALAMRINGDKAAFYNCRFYGFQDTLCDDRGNHFFKNCYIEGTYDFIFGR 223
Query: 742 GRSFYEDCRIESVVKEVAVLTAQQRSKSIEGAIDTGFSFKNCSIGGVKGGQIYLGRAWGD 921
G S Y ++ +V + V+ A R + E G+SF +C + GV G IYLGRAW
Sbjct: 224 GASLYLTTQLHAVGDGLRVIAAHNRQSTTE---QNGYSFVHCKVTGV-GTGIYLGRAWMS 279
Query: 922 SSRVVYSYTKMGEEVVPVGWDGWQIAKPESSGIYYGEFKCFGPGADAKKKKRVGWALDLT 1101
+VVYSYT+M V P GW ++ + ++YGE+ C GPG + K KRV D+
Sbjct: 280 HPKVVYSYTEMSSVVNPSGWQENRV-RAHDKTVFYGEYMCTGPG--SHKAKRVAHTQDID 336
Query: 1102 EAQAKPFVGTHYVLGDTWIQPPP 1170
+A F+ Y+ G W+ PPP
Sbjct: 337 NKEASQFLTLGYIKGSKWLLPPP 359
>sptr|Q9LY19|Q9LY19 Pectin methyl-esterase-like protein.
Length = 342
Score = 220 bits (560), Expect = 4e-56
Identities = 124/326 (38%), Positives = 182/326 (55%)
Frame = +1
Query: 193 KEPLDKKLSEAEKKKVTYVVDPSGKGDYTNITAALEDIPVSNTKRVILDLKPGAQFREKL 372
++ LD L AE V+P G G++ +T A++ +P NTKRVI+ + G ++REK+
Sbjct: 31 RKGLDPALVAAEAAPRIINVNPKG-GEFKTLTDAIKSVPAGNTKRVIIKMAHG-EYREKV 88
Query: 373 FLNISKPFITFRSDPKKPAVVVWNDTAATNGKDGKPVGTVGSATLAVESDYFTAYGVVFR 552
++ +KPFIT P V+ ++ TAA GTV SA+L + SDYF A +V +
Sbjct: 89 TIDRNKPFITLMGQPNAMPVITYDGTAAK-------YGTVDSASLIILSDYFMAVNIVVK 141
Query: 553 NDAPLAKPGAKGGQAVAVRLFGTKTQIYNCTIDGGQDTLYDHKGLHYFKGCLIRGSVDFI 732
N AP KG QA+++R+ G YNC G QDT+ D G H+FK C + G+ DFI
Sbjct: 142 NTAPAPDGKTKGAQALSMRISGNFAAFYNCKFYGFQDTICDDTGNHFFKDCYVEGTFDFI 201
Query: 733 FGFGRSFYEDCRIESVVKEVAVLTAQQRSKSIEGAIDTGFSFKNCSIGGVKGGQIYLGRA 912
FG G S Y ++ V + V+ A + E +G+SF +C + G GG IYLGRA
Sbjct: 202 FGSGTSMYLGTQLHVVGDGIRVIAAHAGKSAEE---KSGYSFVHCKVTGT-GGGIYLGRA 257
Query: 913 WGDSSRVVYSYTKMGEEVVPVGWDGWQIAKPESSGIYYGEFKCFGPGADAKKKKRVGWAL 1092
W +VVY+YT+M V P GW + + + ++YGE+KC GPG + K KRV +
Sbjct: 258 WMSHPKVVYAYTEMTSVVNPTGWQENKTPAHDKT-VFYGEYKCSGPG--SHKAKRVPFTQ 314
Query: 1093 DLTEAQAKPFVGTHYVLGDTWIQPPP 1170
D+ + +A F+ Y+ G W+ PPP
Sbjct: 315 DIDDKEANCFLSLGYIQGSKWLLPPP 340
>sptr|Q9ZQA3|Q9ZQA3 Putative pectinesterase.
Length = 407
Score = 218 bits (554), Expect = 2e-55
Identities = 122/327 (37%), Positives = 183/327 (55%), Gaps = 5/327 (1%)
Frame = +1
Query: 196 EPLDKKLSEAEKKKVTYVVDPSGKGDYTNITAALEDIPVSNTKRVILDLKPGAQFREKLF 375
E +L K + VD G G+++N+ +A++ +P ++ + ++ + G+ +REK+
Sbjct: 75 EKWTSRLRHQYKTSLVLTVDLHGCGNFSNVQSAIDVVPDLSSSKTLIIVNSGS-YREKVT 133
Query: 376 LNISKPFITFRSDPKKPAVVVWNDTAATNGKDGKPVGTVGSATLAVESDYFTAYGVVFRN 555
+N +K + + + + WNDTA + G T S + V + FTAY + F+N
Sbjct: 134 VNENKTNLVIQGRGYQNTSIEWNDTAKSAGN------TADSFSFVVFAANFTAYNISFKN 187
Query: 556 DAPLAKPGAKGGQAVAVRLFGTKTQIYNCTIDGGQDTLYDHKGLHYFKGCLIRGSVDFIF 735
+AP PG QAVA+R+ G + Y C G QDTL D KG H+FK C I+GS+DFIF
Sbjct: 188 NAPEPDPGEADAQAVALRIEGDQAAFYGCGFYGAQDTLLDDKGRHFFKECFIQGSIDFIF 247
Query: 736 GFGRSFYEDCRIESVVK-----EVAVLTAQQRSKSIEGAIDTGFSFKNCSIGGVKGGQIY 900
G GRS Y+DC I S+ K +TAQ R E +GFSF NC I G G+I
Sbjct: 248 GNGRSLYQDCTINSIAKGNTSGVTGSITAQGRQSEDE---QSGFSFVNCKIDG--SGEIL 302
Query: 901 LGRAWGDSSRVVYSYTKMGEEVVPVGWDGWQIAKPESSGIYYGEFKCFGPGADAKKKKRV 1080
LGRAWG + VV+S T M + P GW+ W + E + + +GE KC+GPGAD K+RV
Sbjct: 303 LGRAWGAYATVVFSNTYMSGIITPEGWNNWGDSTKEKT-VTFGEHKCYGPGAD--YKERV 359
Query: 1081 GWALDLTEAQAKPFVGTHYVLGDTWIQ 1161
+ LT+++A F+ ++ GD W++
Sbjct: 360 LFGKQLTDSEASSFIDVSFIDGDEWLR 386
>sptr|Q9CAS7|Q9CAS7 Putative pectin methylesterase.
Length = 338
Score = 217 bits (553), Expect = 3e-55
Identities = 116/301 (38%), Positives = 172/301 (57%)
Frame = +1
Query: 268 GDYTNITAALEDIPVSNTKRVILDLKPGAQFREKLFLNISKPFITFRSDPKKPAVVVWND 447
G++ +T A++ +P NTKRVI+ + PG +++EK+ ++ +KPFIT P V+ ++
Sbjct: 51 GEFKTLTDAIKSVPAGNTKRVIIKMAPG-EYKEKVTIDRNKPFITLMGQPNAMPVITYDG 109
Query: 448 TAATNGKDGKPVGTVGSATLAVESDYFTAYGVVFRNDAPLAKPGAKGGQAVAVRLFGTKT 627
TAA GTV SA+L + SDYF A +V +N AP KG QA+++R+ G
Sbjct: 110 TAAK-------YGTVDSASLIILSDYFMAVNIVVKNTAPAPDGKTKGAQALSMRISGNFA 162
Query: 628 QIYNCTIDGGQDTLYDHKGLHYFKGCLIRGSVDFIFGFGRSFYEDCRIESVVKEVAVLTA 807
YNC G QDT+ D G H+FK C + G+ DFIFG G S Y ++ V + V+ A
Sbjct: 163 AFYNCKFYGFQDTICDDTGNHFFKDCYVEGTFDFIFGSGTSMYLGTQLHVVGDGIRVIAA 222
Query: 808 QQRSKSIEGAIDTGFSFKNCSIGGVKGGQIYLGRAWGDSSRVVYSYTKMGEEVVPVGWDG 987
+ E +G+SF +C + G GG IYLGRAW +VVY+YT+M V P GW
Sbjct: 223 HAGKSAEE---KSGYSFVHCKVTGT-GGGIYLGRAWMSHPKVVYAYTEMTSVVNPTGWQE 278
Query: 988 WQIAKPESSGIYYGEFKCFGPGADAKKKKRVGWALDLTEAQAKPFVGTHYVLGDTWIQPP 1167
+ + + ++YGE+KC GPG + K KRV + D+ + +A F+ Y+ G W+ PP
Sbjct: 279 NKTPAHDKT-VFYGEYKCSGPG--SHKAKRVPFTQDIDDKEANRFLSLGYIQGSKWLLPP 335
Query: 1168 P 1170
P
Sbjct: 336 P 336
>sptr|Q84WQ3|Q84WQ3 Putative pectinesterase family protein.
Length = 407
Score = 215 bits (547), Expect = 1e-54
Identities = 121/327 (37%), Positives = 182/327 (55%), Gaps = 5/327 (1%)
Frame = +1
Query: 196 EPLDKKLSEAEKKKVTYVVDPSGKGDYTNITAALEDIPVSNTKRVILDLKPGAQFREKLF 375
E +L K + VD G G+++N+ +A++ +P ++ + ++ + G+ +REK+
Sbjct: 75 EKWTSRLRHQYKTSLVLTVDLHGCGNFSNVQSAIDVVPDLSSSKTLIIVNSGS-YREKVT 133
Query: 376 LNISKPFITFRSDPKKPAVVVWNDTAATNGKDGKPVGTVGSATLAVESDYFTAYGVVFRN 555
+N +K + + + + WNDTA + G T S + V + FTAY + F+N
Sbjct: 134 VNENKTNLVIQGRGYQNTSIEWNDTAKSAGN------TADSFSFVVFAANFTAYNISFKN 187
Query: 556 DAPLAKPGAKGGQAVAVRLFGTKTQIYNCTIDGGQDTLYDHKGLHYFKGCLIRGSVDFIF 735
+AP PG QAVA+R+ G + Y C G QDTL D KG H+FK C I+GS+ FIF
Sbjct: 188 NAPEPDPGEADAQAVALRIEGDQAAFYGCGFYGAQDTLLDDKGRHFFKECFIQGSIGFIF 247
Query: 736 GFGRSFYEDCRIESVVK-----EVAVLTAQQRSKSIEGAIDTGFSFKNCSIGGVKGGQIY 900
G GRS Y+DC I S+ K +TAQ R E +GFSF NC I G G+I
Sbjct: 248 GNGRSLYQDCTINSIAKGNTSGVTGSITAQGRQSEDE---QSGFSFVNCKIDG--SGEIL 302
Query: 901 LGRAWGDSSRVVYSYTKMGEEVVPVGWDGWQIAKPESSGIYYGEFKCFGPGADAKKKKRV 1080
LGRAWG + VV+S T M + P GW+ W + E + + +GE KC+GPGAD K+RV
Sbjct: 303 LGRAWGAYATVVFSNTYMSGIITPEGWNNWGDSTKEKT-VTFGEHKCYGPGAD--YKERV 359
Query: 1081 GWALDLTEAQAKPFVGTHYVLGDTWIQ 1161
+ LT+++A F+ ++ GD W++
Sbjct: 360 LFGKQLTDSEASSFIDVSFIDGDEWLR 386
>sptr|Q9FGK7|Q9FGK7 Pectin methylesterase-like.
Length = 359
Score = 214 bits (545), Expect = 2e-54
Identities = 116/305 (38%), Positives = 165/305 (54%)
Frame = +1
Query: 250 VDPSGKGDYTNITAALEDIPVSNTKRVILDLKPGAQFREKLFLNISKPFITFRSDPKKPA 429
V +G + ++ A++ IP +N K + + + PG EK+ + +KP+ITF+ +
Sbjct: 63 VSLNGHAQFRSVQDAVDSIPKNNNKSITIKIAPGL---EKVVVPATKPYITFKGAGRDVT 119
Query: 430 VVVWNDTAATNGKDGKPVGTVGSATLAVESDYFTAYGVVFRNDAPLAKPGAKGGQAVAVR 609
+ W+D A+ G +G+ + T +A++ V ++YFTA + F N AP PG +G QAVA R
Sbjct: 120 AIEWHDRASDLGANGQQLRTYQTASVTVYANYFTARNISFTNTAPAPLPGMQGWQAVAFR 179
Query: 610 LFGTKTQIYNCTIDGGQDTLYDHKGLHYFKGCLIRGSVDFIFGFGRSFYEDCRIESVVKE 789
+ G K C G QDTL D G HYFK C I GS+DFIFG GRS Y+DC + S+
Sbjct: 180 ISGDKAFFSGCGFYGAQDTLCDDAGRHYFKECYIEGSIDFIFGNGRSMYKDCELHSIASR 239
Query: 790 VAVLTAQQRSKSIEGAIDTGFSFKNCSIGGVKGGQIYLGRAWGDSSRVVYSYTKMGEEVV 969
+ A R+ E TGF+F C + G G +Y+GRA G SR+VY+YT V
Sbjct: 240 FGSIAAHGRTCPEE---KTGFAFVGCRVTGT--GPLYVGRAMGQYSRIVYAYTYFDALVA 294
Query: 970 PVGWDGWQIAKPESSGIYYGEFKCFGPGADAKKKKRVGWALDLTEAQAKPFVGTHYVLGD 1149
GWD W K ++G + C+GPGA A + V WA L A PF+ +V G
Sbjct: 295 HGGWDDWD-HKSNKRTAFFGVYNCYGPGAAA--TRGVSWARALDYESAHPFIAKSFVNGR 351
Query: 1150 TWIQP 1164
WI P
Sbjct: 352 HWIAP 356
>sptr|O23038|O23038 YUP8H12.7 protein.
Length = 391
Score = 202 bits (515), Expect = 7e-51
Identities = 116/312 (37%), Positives = 173/312 (55%), Gaps = 8/312 (2%)
Frame = +1
Query: 250 VDPSGKGDYTNITAALEDIPVSNTKRVILDLKPGAQFREKLFLNISKPFITFRSDPKKPA 429
VD +G ++T + +A++ + + +R ++ + G EK+ + +KP IT +
Sbjct: 93 VDKNGCCNFTTVQSAVDAVGNFSQRRNVIWINSGI---EKVVIPKTKPNITLQGQGFDIT 149
Query: 430 VVVWNDTAATNGKDGKPVGTVGSATLAVESDYFTAYGVVFRNDAPLAKPGAKGGQAVAVR 609
+ WNDTA + GT AT+ V F A + F N AP+ KPG G QAVA+R
Sbjct: 150 AIAWNDTAYSAN------GTFYCATVQVFGSQFVAKNISFMNVAPIPKPGDVGAQAVAIR 203
Query: 610 LFGTKTQIYNCTIDGGQDTLYDHKGLHYFKGCLIRGSVDFIFGFGRSFYEDCRIESVVKE 789
+ G ++ C G QDTL+D +G HYFK C I+GS+DFIFG +S Y+DCRI S+ +
Sbjct: 204 IAGDESAFVGCGFFGAQDTLHDDRGRHYFKDCYIQGSIDFIFGNAKSLYQDCRIISMANQ 263
Query: 790 VA--------VLTAQQRSKSIEGAIDTGFSFKNCSIGGVKGGQIYLGRAWGDSSRVVYSY 945
++ +TA RS E ++GFSF NC+IGG G ++LGRAW SRVV+
Sbjct: 264 LSPGSKAVNGAVTANGRSSKDE---NSGFSFVNCTIGGT--GHVWLGRAWRPYSRVVFVS 318
Query: 946 TKMGEEVVPVGWDGWQIAKPESSGIYYGEFKCFGPGADAKKKKRVGWALDLTEAQAKPFV 1125
T M + + P GW+ + +++ I+YGE+ C GPGAD KR + L E Q +
Sbjct: 319 TTMTDVIAPEGWNNFNDPSRDAT-IFYGEYNCSGPGAD--MSKRAPYVQKLNETQVALLI 375
Query: 1126 GTHYVLGDTWIQ 1161
T ++ GD W+Q
Sbjct: 376 NTSFIDGDQWLQ 387
>sptr|Q9LSP1|Q9LSP1 Pectinesterase-like protein (Pectinesterase,
putative).
Length = 344
Score = 198 bits (503), Expect = 2e-49
Identities = 109/319 (34%), Positives = 178/319 (55%), Gaps = 4/319 (1%)
Frame = +1
Query: 214 LSEAEKKKVTYVVDPSGKGDYTNITAALEDIPVSNTKRVILDLKPGAQFREKLFLNISKP 393
L+E + +VD GKGDYT++ A++ +PV N+ +I+ ++ G ++E++ + +KP
Sbjct: 36 LTEKIATNRSIIVDIEGKGDYTSVQKAIDAVPVGNSNWIIVHVRKGI-YKERVHIPENKP 94
Query: 394 FITFRSDPKKPAVVVWNDTAATNGKDGKPVGTVGSATLAVESDYFTAYGVVFRNDAPLAK 573
FI R + K V+ + ++ N V SAT VE+++F A+G+ RNDAP+
Sbjct: 95 FIFMRGNGKGKTVIESSQSSVDN---------VASATFKVEANHFVAFGISIRNDAPVGM 145
Query: 574 PGAKGGQAVAVRLFGTKTQIYNCTIDGGQDTLYDHKGLHYFKGCLIRGSVDFIFGFGRSF 753
Q+VA + K Y+C +TL+D+KG HY+ C I+GS+DFIFG S
Sbjct: 146 AFTSENQSVAAFVAADKVAFYHCAFYSLHNTLFDNKGRHYYHECYIQGSIDFIFGRATSI 205
Query: 754 YEDCRI----ESVVKEVAVLTAQQRSKSIEGAIDTGFSFKNCSIGGVKGGQIYLGRAWGD 921
+ +C I + VK +TA R + E TG+ F + G+ ++YLGRA G
Sbjct: 206 FNNCEIFVISDKRVKPYGSITAHHRESAEE---KTGYVFIRGKVYGI--DEVYLGRAKGP 260
Query: 922 SSRVVYSYTKMGEEVVPVGWDGWQIAKPESSGIYYGEFKCFGPGADAKKKKRVGWALDLT 1101
SRV+++ T + + VVP GW W + +Y+GE+KC GPGA+ ++KR WA DLT
Sbjct: 261 YSRVIFAKTYLSKTVVPDGWTNWSY-HGSTQNLYHGEYKCHGPGAE--RQKRSDWAKDLT 317
Query: 1102 EAQAKPFVGTHYVLGDTWI 1158
+ + + F+ ++ G +W+
Sbjct: 318 KQEVESFLSIDFIDGTSWL 336
>sptr|Q9SIJ9|Q9SIJ9 Putative pectinesterase.
Length = 352
Score = 196 bits (499), Expect = 5e-49
Identities = 112/309 (36%), Positives = 169/309 (54%), Gaps = 2/309 (0%)
Frame = +1
Query: 250 VDPSGKGDYTNITAALEDIP--VSNTKRVILDLKPGAQFREKLFLNISKPFITFRSDPKK 423
VD SGKGD++ I A+E IP ++N++ + +KPG +REK+ + KP+IT
Sbjct: 53 VDQSGKGDFSKIQEAIESIPPNLNNSQLYFIWVKPGI-YREKVVIPAEKPYITLSGTQAS 111
Query: 424 PAVVVWNDTAATNGKDGKPVGTVGSATLAVESDYFTAYGVVFRNDAPLAKPGAKGGQAVA 603
++W+D G+D + S TL + + F + +N A G+AVA
Sbjct: 112 NTFLIWSD-----GED-----ILESPTLTIFASDFVCRFLTIQNKFGTA------GRAVA 155
Query: 604 VRLFGTKTQIYNCTIDGGQDTLYDHKGLHYFKGCLIRGSVDFIFGFGRSFYEDCRIESVV 783
+R+ K Y C I QDTL D G HYFK C I G+ DFI G S YE C + S+
Sbjct: 156 LRVAADKAAFYGCVITSYQDTLLDDNGNHYFKNCYIEGATDFICGSASSLYERCHLHSLS 215
Query: 784 KEVAVLTAQQRSKSIEGAIDTGFSFKNCSIGGVKGGQIYLGRAWGDSSRVVYSYTKMGEE 963
+TAQ R+ + E +GF+F C + G G +LGR WG SRVV++Y+
Sbjct: 216 PNNGSITAQMRTSATE---KSGFTFLGCKLTG--SGSTFLGRPWGAYSRVVFAYSFFSNV 270
Query: 964 VVPVGWDGWQIAKPESSGIYYGEFKCFGPGADAKKKKRVGWALDLTEAQAKPFVGTHYVL 1143
V P GW+ W + E++ +YYGE+KC+GPGAD +++RV W+ L++ +A F+ ++
Sbjct: 271 VAPQGWNQWGDSTKENT-VYYGEYKCYGPGAD--REQRVEWSKQLSDEEATVFLSKDFIG 327
Query: 1144 GDTWIQPPP 1170
G W++P P
Sbjct: 328 GKDWLRPAP 336
>sptr|Q8RYM8|Q8RYM8 Pectin methylesterase-like protein.
Length = 293
Score = 194 bits (494), Expect = 2e-48
Identities = 111/314 (35%), Positives = 168/314 (53%), Gaps = 6/314 (1%)
Frame = +1
Query: 238 VTYVVDPSGKGDYTNITAALEDIPVSNTKRVILDLKPGAQFR----EKLFLNISKPFITF 405
V VD SGKGD+ I A++ P +++ R ++ +KPG R EK+ + KP++T
Sbjct: 4 VVVTVDQSGKGDHRRIQDAIDAAPANDSSRTVIRIKPGVYRRVGNQEKVV--VDKPYVTL 61
Query: 406 RSDPKKPAVVVWNDTAATNGKDGKPVGTVGSATLAVESDYFTAYGVVFRNDAPLAKPGAK 585
V+ WN++ ++ S T++V + F A + F+N + P
Sbjct: 62 TGTSATSTVIAWNESWVSDE----------SPTVSVLASDFVAKRLTFQNTFGDSAP--- 108
Query: 586 GGQAVAVRLFGTKTQIYNCTIDGGQDTLYDHKGLHYFKGCLIRGSVDFIFGFGRSFYEDC 765
AVAVR+ G + Y C QDTL D G HY++GC ++G+ DFIFG GR+ ++ C
Sbjct: 109 ---AVAVRVAGDRAAFYGCRFVSFQDTLLDETGRHYYRGCYVQGATDFIFGNGRALFDKC 165
Query: 766 RIESVVKEVA--VLTAQQRSKSIEGAIDTGFSFKNCSIGGVKGGQIYLGRAWGDSSRVVY 939
+ S + A TAQQRS E +TG+SF C + G+ G LGR WG SRVV+
Sbjct: 166 HLHSTSPDGAGGAFTAQQRSSESE---ETGYSFVGCKLTGLGAGTSILGRPWGPYSRVVF 222
Query: 940 SYTKMGEEVVPVGWDGWQIAKPESSGIYYGEFKCFGPGADAKKKKRVGWALDLTEAQAKP 1119
+ T M V P GWD W + + +YG+++C+G G +K RV W+ DLT+A+A P
Sbjct: 223 ALTYMSSTVRPQGWDDWGDPSNQRTA-FYGQYQCYGDG--SKTDGRVAWSHDLTQAEAAP 279
Query: 1120 FVGTHYVLGDTWIQ 1161
F+ +V G W++
Sbjct: 280 FITKAWVDGQQWLR 293
>sptr|Q9ZQA4|Q9ZQA4 Putative pectinesterase.
Length = 333
Score = 194 bits (492), Expect = 3e-48
Identities = 116/319 (36%), Positives = 174/319 (54%), Gaps = 4/319 (1%)
Frame = +1
Query: 214 LSEAEKKKVTYVVDPS--GKGDYTNITAALEDIPVSNTKRVILDLKPGAQFREKLFLNIS 387
LS+ K T V+ S G G + + A+ D + +++ L L +RE+ ++ +
Sbjct: 30 LSKFPTKGFTMVLKVSLNGCGRFKRVQDAI-DASIGSSQSKTLILIDFGIYRERFIVHEN 88
Query: 388 KPFITFRSDPKKPAVVVWNDTAATNGKDGKPVGTVGSATLAVESDYFTAYGVVFRNDAPL 567
K + + + WN+T A++ GT S ++AV + FTAY + F+N AP
Sbjct: 89 KNNLVVQGMGYSRTSIEWNNTTASSN------GTFSSFSVAVFGEKFTAYNISFKNTAPA 142
Query: 568 AKPGAKGGQAVAVRLFGTKTQIYNCTIDGGQDTLYDHKGLHYFKGCLIRGSVDFIFGFGR 747
PGA QAVA+++ G K Y C G QDTL D +G H+FKGC I GS+DFIFG GR
Sbjct: 143 PNPGAVDAQAVALKVVGDKAAFYGCGFYGNQDTLLDQEGRHFFKGCFIEGSIDFIFGNGR 202
Query: 748 SFYEDCRIESVVKE--VAVLTAQQRSKSIEGAIDTGFSFKNCSIGGVKGGQIYLGRAWGD 921
S YEDC + S+ KE + +TA + + TGF F NC I G +++LGRAW
Sbjct: 203 SLYEDCTLHSIAKENTIGCITANGKDTLKD---RTGFVFVNCKITG--SARVWLGRAWRP 257
Query: 922 SSRVVYSYTKMGEEVVPVGWDGWQIAKPESSGIYYGEFKCFGPGADAKKKKRVGWALDLT 1101
+RV++S T M V GW+ K + + +YYGE +C+GPGA+ KRV +A L+
Sbjct: 258 YARVIFSKTYMSRVVSLDGWNDMGDPKTQRT-VYYGEHRCYGPGAN--HSKRVTYAKLLS 314
Query: 1102 EAQAKPFVGTHYVLGDTWI 1158
+ +A PF ++ G+ W+
Sbjct: 315 DVEAAPFTNISFIDGEEWL 333
>sptr|Q9LVQ0|Q9LVQ0 Pectin methylesterase-like protein (Putative
pectinesterase).
Length = 317
Score = 189 bits (480), Expect = 8e-47
Identities = 115/302 (38%), Positives = 158/302 (52%), Gaps = 5/302 (1%)
Frame = +1
Query: 250 VDPSGKGDYTNITAALEDIPVSNTKRVILDLKPGAQFREKLFLNISKPFITFRSDPKKPA 429
V G GDY ++ A++ +P+ NT R ++ L PG +R+ +++ K FITF +
Sbjct: 9 VSQDGSGDYCSVQDAIDSVPLGNTCRTVIRLSPGI-YRQPVYVPKRKNFITFAGISPEIT 67
Query: 430 VVVWNDTAAT----NGKDGKPVGTVGSATLAVESDYFTAYGVVFRNDAPLAKPGAKGGQA 597
V+ WN+TA+ GT G ++ VE + F A + F N AP GQA
Sbjct: 68 VLTWNNTASKIEHHQASRVIGTGTFGCGSVIVEGEDFIAENITFENSAPEGS-----GQA 122
Query: 598 VAVRLFGTKTQIYNCTIDGGQDTLYDHKGLHYFKGCLIRGSVDFIFGFGRSFYEDCRIES 777
VA+R+ + YNC G QDTLY H G Y K C I GSVDFIFG + E C I
Sbjct: 123 VAIRVTADRCAFYNCRFLGWQDTLYLHHGKQYLKDCYIEGSVDFIFGNSTALLEHCHIH- 181
Query: 778 VVKEVAVLTAQQRSKSIEGAIDTGFSFKNCSI-GGVKGGQIYLGRAWGDSSRVVYSYTKM 954
K +TAQ R S E TG+ F C I G + G +YLGR WG RVV +YT M
Sbjct: 182 -CKSQGFITAQSRKSSQE---STGYVFLRCVITGNGQSGYMYLGRPWGPFGRVVLAYTYM 237
Query: 955 GEEVVPVGWDGWQIAKPESSGIYYGEFKCFGPGADAKKKKRVGWALDLTEAQAKPFVGTH 1134
+ VGW W A+ E S +Y E++CFGPG+ + +RV W+ +L + +A FV
Sbjct: 238 DACIRNVGWHNWGNAENERSACFY-EYRCFGPGSCS--SERVPWSRELMDDEAGHFVHHS 294
Query: 1135 YV 1140
+V
Sbjct: 295 FV 296
>sptr|Q7XEU2|Q7XEU2 Putative pectin methylesterase.
Length = 336
Score = 166 bits (421), Expect = 6e-40
Identities = 98/300 (32%), Positives = 152/300 (50%), Gaps = 7/300 (2%)
Frame = +1
Query: 262 GKGDYTNITAALEDIPVSNTKRVILDLKPGAQFREKLFLNISKPFITFRSDPKKP--AVV 435
G+ + + AA++ +PV N R ++ L PG +RE +++ +K +T + P V+
Sbjct: 30 GEAVFATVQAAVDAVPVGNRVRTVIRLAPGT-YREPVYVAKAKNLVTLSGEAGSPEATVI 88
Query: 436 VWNDTAAT--NGKDGKPVGT--VGSATLAVESDYFTAYGVVFRNDAPLAKPGAKGGQAVA 603
W++TA + + + +GT G T+ VE + F A + F N AP GQAVA
Sbjct: 89 TWDNTATRIKHSQSSRVIGTGTFGCGTIIVEGEDFIAENITFENSAPQGS-----GQAVA 143
Query: 604 VRLFGTKTQIYNCTIDGGQDTLYDHKGLHYFKGCLIRGSVDFIFGFGRSFYEDCRIESVV 783
+R+ + YNC G QDTLY H G Y + C I G+ DFIFG + E C I
Sbjct: 144 LRVTADRCAFYNCRFLGWQDTLYLHYGKQYLRDCYIEGNCDFIFGNSIALLEHCHIH--C 201
Query: 784 KEVAVLTAQQRSKSIEGAIDTGFSFKNCSI-GGVKGGQIYLGRAWGDSSRVVYSYTKMGE 960
K +TA R S E TG+ F C I G + G ++LGR WG RVV+++T M
Sbjct: 202 KSAGYITAHSRKSSSE---TTGYVFLRCIITGNGEAGYMFLGRPWGPFGRVVFAHTFMDR 258
Query: 961 EVVPVGWDGWQIAKPESSGIYYGEFKCFGPGADAKKKKRVGWALDLTEAQAKPFVGTHYV 1140
+ P GW W ++ E + ++ E++C GPG + RV W L + + + F+ ++
Sbjct: 259 CIKPAGWHNWDRSENERTACFF-EYRCSGPG--FRPSNRVAWCRQLLDVEVENFLSHSFI 315
>sptr|Q948H9|Q948H9 Putative pectin methylesterase.
Length = 336
Score = 166 bits (421), Expect = 6e-40
Identities = 98/300 (32%), Positives = 152/300 (50%), Gaps = 7/300 (2%)
Frame = +1
Query: 262 GKGDYTNITAALEDIPVSNTKRVILDLKPGAQFREKLFLNISKPFITFRSDPKKP--AVV 435
G+ + + AA++ +PV N R ++ L PG +RE +++ +K +T + P V+
Sbjct: 30 GEAVFATVQAAVDAVPVGNRVRTVIRLAPGT-YREPVYVAKAKNLVTLSGEAGSPEATVI 88
Query: 436 VWNDTAAT--NGKDGKPVGT--VGSATLAVESDYFTAYGVVFRNDAPLAKPGAKGGQAVA 603
W++TA + + + +GT G T+ VE + F A + F N AP GQAVA
Sbjct: 89 TWDNTATRIKHSQSSRVIGTGTFGCGTIIVEGEDFIAENITFENSAPQGS-----GQAVA 143
Query: 604 VRLFGTKTQIYNCTIDGGQDTLYDHKGLHYFKGCLIRGSVDFIFGFGRSFYEDCRIESVV 783
+R+ + YNC G QDTLY H G Y + C I G+ DFIFG + E C I
Sbjct: 144 LRVTADRCAFYNCRFLGWQDTLYLHYGKQYLRDCYIEGNCDFIFGNSIALLEHCHIH--C 201
Query: 784 KEVAVLTAQQRSKSIEGAIDTGFSFKNCSI-GGVKGGQIYLGRAWGDSSRVVYSYTKMGE 960
K +TA R S E TG+ F C I G + G ++LGR WG RVV+++T M
Sbjct: 202 KSAGYITAHSRKSSSE---TTGYVFLRCIITGNGEAGYMFLGRPWGPFGRVVFAHTFMDR 258
Query: 961 EVVPVGWDGWQIAKPESSGIYYGEFKCFGPGADAKKKKRVGWALDLTEAQAKPFVGTHYV 1140
+ P GW W ++ E + ++ E++C GPG + RV W L + + + F+ ++
Sbjct: 259 CIKPAGWHNWDRSENERTACFF-EYRCSGPG--FRPSNRVAWCRQLLDVEVENFLSHSFI 315
>sptr|Q93VX7|Q93VX7 Putative pectin methylesterase.
Length = 335
Score = 153 bits (386), Expect = 7e-36
Identities = 100/310 (32%), Positives = 149/310 (48%), Gaps = 2/310 (0%)
Frame = +1
Query: 250 VDPSGKG-DYTNITAALEDIPVSNTKRVILDLKPGAQFREKLFLNISKPFITFRSDPKKP 426
V G G D+T I A+ +P +N + + + + G ++EK+ + +K FI + ++
Sbjct: 44 VSKKGSGADFTRIQDAINSVPFANRRWIRIHIAAGV-YKEKVSIPANKSFILLEGEGRQQ 102
Query: 427 AVVVWNDTAATNGKDGKPVGTVGSATLAVESDYFTAYGVVFRNDAPLAKPGAKGGQAVAV 606
+ W D A G D GT S T A + F A + F+N P AVA
Sbjct: 103 TSIEWADHAGGGGGDS---GTADSPTFASYAADFMARDITFKNTYGRMAP------AVAA 153
Query: 607 RLFGTKTQIYNCTIDGGQDTLYDHKGLHYFKGCLIRGSVDFIFGFGRSFYEDCRIESVVK 786
+ G ++ Y C G QDTL D G HY++ C + G+VDFIFG +S + C I +
Sbjct: 154 LVAGDRSAFYRCGFVGLQDTLSDLLGRHYYERCYVEGAVDFIFGEAQSIFHRCHISTAAA 213
Query: 787 EV-AVLTAQQRSKSIEGAIDTGFSFKNCSIGGVKGGQIYLGRAWGDSSRVVYSYTKMGEE 963
+TAQ RS + + +GF F +C++GG YLGRAW +RVV+ T M
Sbjct: 214 AAPGFITAQGRSSASDA---SGFVFTSCTVGG--AAPAYLGRAWRAYARVVFYRTAMSAA 268
Query: 964 VVPVGWDGWQIAKPESSGIYYGEFKCFGPGADAKKKKRVGWALDLTEAQAKPFVGTHYVL 1143
VV +GWD W K + + E C GPG++ + RV W L+ + V YV
Sbjct: 269 VVGLGWDAWDY-KGKEETLEMVESGCTGPGSN--RTGRVPWEKTLSGEELAKLVDISYVS 325
Query: 1144 GDTWIQPPPK 1173
D W+ P+
Sbjct: 326 RDGWLAAQPR 335
>sptr|Q9FJ21|Q9FJ21 Pectin methylesterase (EC 3.1.1.11)
(AT5g49180/K21P3_5) (Pectinesterase).
Length = 571
Score = 153 bits (386), Expect = 7e-36
Identities = 108/321 (33%), Positives = 157/321 (48%), Gaps = 11/321 (3%)
Frame = +1
Query: 235 KVTYVVDPSGKGDYTNITAALEDIPVSNTKRVILDLKPGAQFREKLFLNISKPFITFRSD 414
K VV G G Y I AL +P +N K ++ +K G + EK+ + +TF D
Sbjct: 257 KANVVVAHDGSGQYKTINEALNAVPKANQKPFVIYIKQGV-YNEKVDVTKKMTHVTFIGD 315
Query: 415 -PKKPAVVVWNDTAATNGKDGKPVGTVGSATLAVESDYFTAYGVVFRNDAPLAKPGAKGG 591
P K + T + N GK V T +AT+A+ D FTA + F N A G +G
Sbjct: 316 GPTKTKI-----TGSLNYYIGK-VKTYLTATVAINGDNFTAKNIGFENTA-----GPEGH 364
Query: 592 QAVAVRLFGTKTQIYNCTIDGGQDTLYDHKGLHYFKGCLIRGSVDFIFGFGRSFYEDCRI 771
QAVA+R+ YNC IDG QDTLY H +F+ C + G+VDFIFG G ++C I
Sbjct: 365 QAVALRVSADLAVFYNCQIDGYQDTLYVHSHRQFFRDCTVSGTVDFIFGDGIVVLQNCNI 424
Query: 772 ---ESVVKEVAVLTAQQRSKSIEGAIDTGFSFKNCSIGG------VKG-GQIYLGRAWGD 921
+ + + ++TAQ RS E TG +NC I G VK + YLGR W +
Sbjct: 425 VVRKPMKSQSCMITAQGRSDKRE---STGLVLQNCHITGEPAYIPVKSINKAYLGRPWKE 481
Query: 922 SSRVVYSYTKMGEEVVPVGWDGWQIAKPESSGIYYGEFKCFGPGADAKKKKRVGWALDLT 1101
SR + T + + + P GW W + +YY E++ GPG++ ++ + L+
Sbjct: 482 FSRTIIMGTTIDDVIDPAGWLPWN-GDFALNTLYYAEYENNGPGSNQAQRVKWPGIKKLS 540
Query: 1102 EAQAKPFVGTHYVLGDTWIQP 1164
QA F ++ G+ WI P
Sbjct: 541 PKQALRFTPARFLRGNLWIPP 561
>sptr|O64479|O64479 Putative pectinesterase.
Length = 339
Score = 151 bits (381), Expect = 2e-35
Identities = 94/316 (29%), Positives = 151/316 (47%), Gaps = 6/316 (1%)
Frame = +1
Query: 241 TYVVDPSGKGDYTNITAALEDIPVSNTKRVILDLKPGAQFREKLFLNISKPFITFRSDPK 420
T +V+P+ + + +A++ IP+ N + + + G + EK+ + K +I +
Sbjct: 40 TIIVNPNDARYFKTVQSAIDSIPLQNQDWIRILISNGI-YSEKVTIPRGKGYIYMQGGGI 98
Query: 421 KPAVVVWNDTAATNGKDGKPVGTVGSATLAVESDYFTAYGVVFRNDAPLAKPGAKGGQAV 600
+ ++ + D TN SAT G+ F+N +A + AV
Sbjct: 99 EKTIIAYGDHQLTNT----------SATFTSYPSNIIITGITFKNKYNIASSSSPTKPAV 148
Query: 601 AVRLFGTKTQIYNCTIDGGQDTLYDHKGLHYFKGCLIRGSVDFIFGFGRSFYEDCRIESV 780
A + G K I + + DG QDTLYD G HY+K C+I G +DFIFG +S +E C ++
Sbjct: 149 AAMMLGDKYAIIDSSFDGFQDTLYDDYGRHYYKRCVISGGIDFIFGGAQSIFEGCTLKLR 208
Query: 781 V------KEVAVLTAQQRSKSIEGAIDTGFSFKNCSIGGVKGGQIYLGRAWGDSSRVVYS 942
V + +TAQ R + GF FK+C++ G G+ LGRAW SRV++
Sbjct: 209 VGIYPPNEVYGTITAQGRDSPTDKG---GFVFKDCTVMG--SGKALLGRAWKSYSRVIFY 263
Query: 943 YTKMGEEVVPVGWDGWQIAKPESSGIYYGEFKCFGPGADAKKKKRVGWALDLTEAQAKPF 1122
+ + ++P+GWD W+ AK + I + EF C G GAD KRV W +E F
Sbjct: 264 RSMFSDNILPIGWDAWK-AKGQEGHITFVEFGCTGVGADT--SKRVPWLTKASEKDVLQF 320
Query: 1123 VGTHYVLGDTWIQPPP 1170
++ + W+ P
Sbjct: 321 TNLTFIDEEGWLSRLP 336
>sptr|Q8GXA1|Q8GXA1 Putative pectin methylesterase.
Length = 568
Score = 150 bits (378), Expect = 6e-35
Identities = 103/320 (32%), Positives = 153/320 (47%), Gaps = 12/320 (3%)
Frame = +1
Query: 235 KVTYVVDPSGKGDYTNITAALEDIPVSNTKRVILDLKPGAQFREKLFLNISKPFITFRSD 414
K VV G G + IT AL +P N I+ +K G ++EK+ + P +TF D
Sbjct: 255 KANAVVAQDGTGQFKTITDALNAVPKGNKVPFIIHIKEGI-YKEKVTVTKKMPHVTFIGD 313
Query: 415 PKKPAVVVWNDTAATNGKDGKPVGTVGSATLAVESDYFTAYGVVFRNDAPLAKPGAKGGQ 594
++ T + N GK V T +AT+ +E D+FTA + N A G +GGQ
Sbjct: 314 GPNKTLI----TGSLNFGIGK-VKTFLTATITIEGDHFTAKNIGIENTA-----GPEGGQ 363
Query: 595 AVAVRLFGTKTQIYNCTIDGGQDTLYDHKGLHYFKGCLIRGSVDFIFGFGRSFYEDCRIE 774
AVA+R+ ++C IDG QDTLY H +++ C + G+VDFIFG + ++C+I
Sbjct: 364 AVALRVSADYAVFHSCQIDGHQDTLYVHSHRQFYRDCTVSGTVDFIFGDAKCILQNCKI- 422
Query: 775 SVVK-----EVAVLTAQQRSKSIEGAIDTGFSFKNCSIGGVKG-------GQIYLGRAWG 918
VV+ + ++TAQ RS E TG C I G + YLGR W
Sbjct: 423 -VVRKPNKGQTCMVTAQGRSNVRE---STGLVLHGCHITGDPAYIPMKSVNKAYLGRPWK 478
Query: 919 DSSRVVYSYTKMGEEVVPVGWDGWQIAKPESSGIYYGEFKCFGPGADAKKKKRVGWALDL 1098
+ SR + T + + + P GW W +YY E GPG++ ++ + L
Sbjct: 479 EFSRTIIMKTTIDDVIDPAGWLPWS-GDFALKTLYYAEHMNTGPGSNQAQRVKWPGIKKL 537
Query: 1099 TEAQAKPFVGTHYVLGDTWI 1158
T A + G ++ GDTWI
Sbjct: 538 TPQDALLYTGDRFLRGDTWI 557
>sptr|Q9M7Y9|Q9M7Y9 Pectin methylesterase, putative (EC 3.1.1.11)
(Pectinesterase).
Length = 562
Score = 150 bits (378), Expect = 6e-35
Identities = 103/320 (32%), Positives = 153/320 (47%), Gaps = 12/320 (3%)
Frame = +1
Query: 235 KVTYVVDPSGKGDYTNITAALEDIPVSNTKRVILDLKPGAQFREKLFLNISKPFITFRSD 414
K VV G G + IT AL +P N I+ +K G ++EK+ + P +TF D
Sbjct: 249 KANAVVAQDGTGQFKTITDALNAVPKGNKVPFIIHIKEGI-YKEKVTVTKKMPHVTFIGD 307
Query: 415 PKKPAVVVWNDTAATNGKDGKPVGTVGSATLAVESDYFTAYGVVFRNDAPLAKPGAKGGQ 594
++ T + N GK V T +AT+ +E D+FTA + N A G +GGQ
Sbjct: 308 GPNKTLI----TGSLNFGIGK-VKTFLTATITIEGDHFTAKNIGIENTA-----GPEGGQ 357
Query: 595 AVAVRLFGTKTQIYNCTIDGGQDTLYDHKGLHYFKGCLIRGSVDFIFGFGRSFYEDCRIE 774
AVA+R+ ++C IDG QDTLY H +++ C + G+VDFIFG + ++C+I
Sbjct: 358 AVALRVSADYAVFHSCQIDGHQDTLYVHSHRQFYRDCTVSGTVDFIFGDAKCILQNCKI- 416
Query: 775 SVVK-----EVAVLTAQQRSKSIEGAIDTGFSFKNCSIGGVKG-------GQIYLGRAWG 918
VV+ + ++TAQ RS E TG C I G + YLGR W
Sbjct: 417 -VVRKPNKGQTCMVTAQGRSNVRE---STGLVLHGCHITGDPAYIPMKSVNKAYLGRPWK 472
Query: 919 DSSRVVYSYTKMGEEVVPVGWDGWQIAKPESSGIYYGEFKCFGPGADAKKKKRVGWALDL 1098
+ SR + T + + + P GW W +YY E GPG++ ++ + L
Sbjct: 473 EFSRTIIMKTTIDDVIDPAGWLPWS-GDFALKTLYYAEHMNTGPGSNQAQRVKWPGIKKL 531
Query: 1099 TEAQAKPFVGTHYVLGDTWI 1158
T A + G ++ GDTWI
Sbjct: 532 TPQDALLYTGDRFLRGDTWI 551
>sptr|Q9LRN4|Q9LRN4 Pectin methylesterase-like protein.
Length = 331
Score = 149 bits (375), Expect = 1e-34
Identities = 96/312 (30%), Positives = 149/312 (47%), Gaps = 5/312 (1%)
Frame = +1
Query: 250 VDPSGKGDYTNITAALEDIPVSNTKRVILDLKPGAQFREKLFLNISKPFITFRSDPKKPA 429
VD SG G++T I A++ +P++N +++K G +REK+ + KPFI K+
Sbjct: 36 VDQSGHGNFTTIQKAIDSVPINNRHWFFINVKAGL-YREKIKIPYEKPFIVLVGAGKRLT 94
Query: 430 VVVWNDTAATNGKDGKPVGTVGSATLAVESDYFTAYGVVFRNDAPLAKPGAKGGQAVAVR 609
V W+D + S T + +D + F AVA
Sbjct: 95 RVEWDDHYSV----------AQSPTFSTLADNTVVKSITFAVRCKGKMNKNPRTPAVAAL 144
Query: 610 LFGTKTQIYNCTIDGGQDTLYDHKGLHYFKGCLIRGSVDFIFGFGRSFYEDCRIESVVKE 789
+ G K+ Y+ G QDTL+D G HYF C I+G+VDFIFG G+S Y+ C I+ + +
Sbjct: 145 IGGDKSAFYSVGFAGIQDTLWDFDGRHYFHRCTIQGAVDFIFGTGQSIYQSCVIQVLGGQ 204
Query: 790 V-----AVLTAQQRSKSIEGAIDTGFSFKNCSIGGVKGGQIYLGRAWGDSSRVVYSYTKM 954
+ +TAQ R+ + GF F NC + G G +LGR W SRV++ + +
Sbjct: 205 LEPGLAGYITAQGRTNPYDA---NGFIFINCLVYGT--GMAFLGRPWRGYSRVIFYNSNL 259
Query: 955 GEEVVPVGWDGWQIAKPESSGIYYGEFKCFGPGADAKKKKRVGWALDLTEAQAKPFVGTH 1134
+ VVP GWD W E+ + + E CFG GA+ +RV W L+E+ +
Sbjct: 260 TDVVVPEGWDAWNFVGHENQ-LVFAEHGCFGSGANI--GRRVKWVKKLSESAIQNLADLS 316
Query: 1135 YVLGDTWIQPPP 1170
++ W++ P
Sbjct: 317 FINRGGWVEDLP 328
>sptr|Q7XAB4|Q7XAB4 Putative pectinesterase.
Length = 345
Score = 148 bits (373), Expect = 2e-34
Identities = 97/312 (31%), Positives = 158/312 (50%), Gaps = 4/312 (1%)
Frame = +1
Query: 235 KVTYVVDPSGKGDYTNITAALEDIPVSNTKRVILDLKPGAQFREKLFLNISKPFITFRSD 414
K T +V P + ++ + +A++ +PV NT+ VI+ L+ G +REK+ + +KPFI R +
Sbjct: 50 KRTLIVGP--EDEFKTVQSAIDAVPVGNTEWVIVHLRSGI-YREKVMIPETKPFIFVRGN 106
Query: 415 PKKPAVVVWNDTAATNGKDGKPVGTVGSATLAVESDYFTAYGVVFRNDAPLAKPGAKGGQ 594
K + ++ N + SA V +D +G+ RN A P +
Sbjct: 107 GKGRTSINHESASSHNAE---------SAAFTVHADNVIVFGLSIRNSARAGLPNVPEVR 157
Query: 595 AVAVRLFGTKTQIYNCTIDGGQDTLYDHKGLHYFKGCLIRGSVDFIFGFGRSFYEDCRI- 771
VA + G K Y+C TL+D G HY++ C I+G++DFIFG G+S ++ I
Sbjct: 158 TVAAMVGGDKIAFYHCAFYSPHHTLFDVAGRHYYESCYIQGNIDFIFGGGQSIFQCPEIF 217
Query: 772 ---ESVVKEVAVLTAQQRSKSIEGAIDTGFSFKNCSIGGVKGGQIYLGRAWGDSSRVVYS 942
+ + +TAQ R K +G +GF F + GV GQ+YLGRA SRV+++
Sbjct: 218 VKPDRRTEIKGSITAQNR-KQEDG---SGFVFIKGKVYGV--GQVYLGRANEAYSRVIFA 271
Query: 943 YTKMGEEVVPVGWDGWQIAKPESSGIYYGEFKCFGPGADAKKKKRVGWALDLTEAQAKPF 1122
T + + + P GW + + + GEF C GPG++A KR W+ LT+ +A F
Sbjct: 272 DTYLSKTINPAGWTSYGYT-GSTDHVMLGEFNCTGPGSEA--TKREPWSRQLTQEEADKF 328
Query: 1123 VGTHYVLGDTWI 1158
+ ++ G W+
Sbjct: 329 INIDFINGKEWL 340
Database: /db/uniprot/tmp/swall
Posted date: Mar 5, 2004 7:45 PM
Number of letters in database: 442,889,342
Number of sequences in database: 1,395,590
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 756,358,788
Number of Sequences: 1395590
Number of extensions: 16553380
Number of successful extensions: 58703
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 53727
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 58038
length of database: 442,889,342
effective HSP length: 126
effective length of database: 267,045,002
effective search space used: 82783950620
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

