BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= 3023585.2.1
(635 letters)
Database: /db/trembl-ebi/tmp/swall
1,288,562 sequences; 411,996,502 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
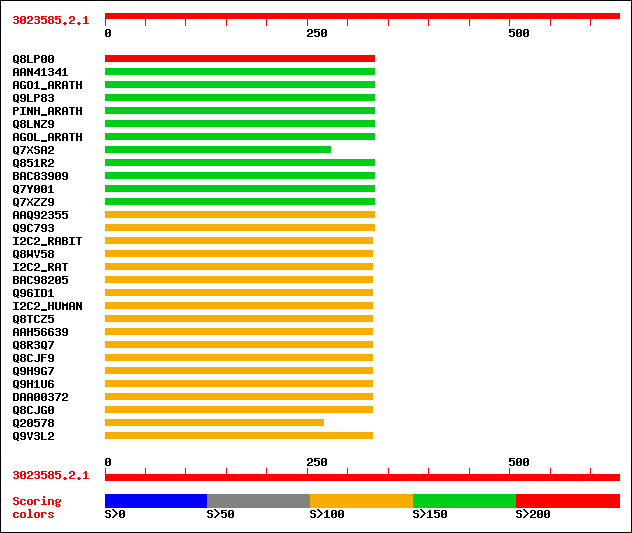
sptr|Q8LP00|Q8LP00 ZLL/PNH homologous protein. 224 5e-58
sptrnew|AAN41341|AAN41341 Putative leaf development protein Argo... 190 1e-47
sw|O04379|AGO1_ARATH Argonaute protein. 190 1e-47
sptr|Q9LP83|Q9LP83 T1N15.2. 190 1e-47
sw|Q9XGW1|PINH_ARATH PINHEAD protein (ZWILLE protein). 189 3e-47
sptr|Q8LNZ9|Q8LNZ9 AGO1 homologous protein. 186 2e-46
sw|Q9SJK3|AGOL_ARATH Argonaute-like protein At2g27880. 177 1e-43
sptr|Q7XSA2|Q7XSA2 OSJNBa0005N02.3 protein. 171 9e-42
sptr|Q851R2|Q851R2 Putative argonaute protein. 170 1e-41
sptrnew|BAC83909|BAC83909 Putative leaf development protein Argo... 166 3e-40
sptr|Q7Y001|Q7Y001 Putative leaf development and shoot apical me... 165 5e-40
sptr|Q7XZZ9|Q7XZZ9 Putative leaf development and shoot apical me... 157 1e-37
sptrnew|AAQ92355|AAQ92355 ZIPPY. 147 1e-34
sptr|Q9C793|Q9C793 Pinhead-like protein. 147 1e-34
sw|O77503|I2C2_RABIT Eukaryotic translation initiation factor 2C... 135 3e-31
sptr|Q8WV58|Q8WV58 Hypothetical protein (Eukaryotic translation ... 135 3e-31
sw|Q9QZ81|I2C2_RAT Eukaryotic translation initiation factor 2C 2... 135 3e-31
sptrnew|BAC98205|BAC98205 MKIAA1567 protein (Fragment). 135 3e-31
sptr|Q96ID1|Q96ID1 Hypothetical protein. 135 3e-31
sw|Q9UKV8|I2C2_HUMAN Eukaryotic translation initiation factor 2C... 135 3e-31
sptr|Q8TCZ5|Q8TCZ5 Eukaryotic initiation factor 2C2. 135 3e-31
sptrnew|AAH56639|AAH56639 Hypothetical protein (Fragment). 135 3e-31
sptr|Q8R3Q7|Q8R3Q7 Similar to eukaryotic translation initiation ... 135 3e-31
sptr|Q8CJF9|Q8CJF9 Piwi/Argonaute family protain meIF2C3. 135 5e-31
sptr|Q9H9G7|Q9H9G7 Hypothetical protein FLJ12765. 135 5e-31
sptr|Q9H1U6|Q9H1U6 DJ665N4.6 (Novel protein) (Fragment). 135 5e-31
sptrnew|DAA00372|DAA00372 Argonaute 3. 135 5e-31
sptr|Q8CJG0|Q8CJG0 Piwi/Argonaute family protain meIF2C2. 134 9e-31
sptr|Q20578|Q20578 F48F7.1 protein. 134 1e-30
sptr|Q9V3L2|Q9V3L2 AGO1 protein. 132 3e-30
>sptr|Q8LP00|Q8LP00 ZLL/PNH homologous protein.
Length = 978
Score = 224 bits (572), Expect = 5e-58
Identities = 108/112 (96%), Positives = 108/112 (96%), Gaps = 1/112 (0%)
Frame = +1
Query: 1 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAY 180
EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAY
Sbjct: 867 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAY 926
Query: 181 YAHLAAFRARFYMEPEMSENQ-TSKSSNGTNGGLVKPLPAVKEKVKRVMFYC 333
YAHLAAFRARFYMEPEMSENQ TSKSS GTNG VKPLPAVKEKVKRVMFYC
Sbjct: 927 YAHLAAFRARFYMEPEMSENQTTSKSSTGTNGTSVKPLPAVKEKVKRVMFYC 978
>sptrnew|AAN41341|AAN41341 Putative leaf development protein
Argonaute.
Length = 1048
Score = 190 bits (483), Expect = 1e-47
Identities = 91/125 (72%), Positives = 100/125 (80%), Gaps = 14/125 (11%)
Frame = +1
Query: 1 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAY 180
EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTAD +Q+LTNNLCYTYARCTRSVS+VPPAY
Sbjct: 924 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADGLQSLTNNLCYTYARCTRSVSIVPPAY 983
Query: 181 YAHLAAFRARFYMEPEMSENQTSKSSNGTNGG--------------LVKPLPAVKEKVKR 318
YAHLAAFRARFYMEPE S++ + S + GG V+PLPA+KE VKR
Sbjct: 984 YAHLAAFRARFYMEPETSDSGSMASGSMARGGGMAGRSTRGPNVNAAVRPLPALKENVKR 1043
Query: 319 VMFYC 333
VMFYC
Sbjct: 1044 VMFYC 1048
>sw|O04379|AGO1_ARATH Argonaute protein.
Length = 1048
Score = 190 bits (483), Expect = 1e-47
Identities = 91/125 (72%), Positives = 100/125 (80%), Gaps = 14/125 (11%)
Frame = +1
Query: 1 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAY 180
EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTAD +Q+LTNNLCYTYARCTRSVS+VPPAY
Sbjct: 924 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADGLQSLTNNLCYTYARCTRSVSIVPPAY 983
Query: 181 YAHLAAFRARFYMEPEMSENQTSKSSNGTNGG--------------LVKPLPAVKEKVKR 318
YAHLAAFRARFYMEPE S++ + S + GG V+PLPA+KE VKR
Sbjct: 984 YAHLAAFRARFYMEPETSDSGSMASGSMARGGGMAGRSTRGPNVNAAVRPLPALKENVKR 1043
Query: 319 VMFYC 333
VMFYC
Sbjct: 1044 VMFYC 1048
>sptr|Q9LP83|Q9LP83 T1N15.2.
Length = 1123
Score = 190 bits (483), Expect = 1e-47
Identities = 91/125 (72%), Positives = 100/125 (80%), Gaps = 14/125 (11%)
Frame = +1
Query: 1 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAY 180
EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTAD +Q+LTNNLCYTYARCTRSVS+VPPAY
Sbjct: 999 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADGLQSLTNNLCYTYARCTRSVSIVPPAY 1058
Query: 181 YAHLAAFRARFYMEPEMSENQTSKSSNGTNGG--------------LVKPLPAVKEKVKR 318
YAHLAAFRARFYMEPE S++ + S + GG V+PLPA+KE VKR
Sbjct: 1059 YAHLAAFRARFYMEPETSDSGSMASGSMARGGGMAGRSTRGPNVNAAVRPLPALKENVKR 1118
Query: 319 VMFYC 333
VMFYC
Sbjct: 1119 VMFYC 1123
>sw|Q9XGW1|PINH_ARATH PINHEAD protein (ZWILLE protein).
Length = 988
Score = 189 bits (479), Expect = 3e-47
Identities = 89/116 (76%), Positives = 97/116 (83%), Gaps = 5/116 (4%)
Frame = +1
Query: 1 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAY 180
EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTAD +Q+LTNNLCYTYARCTRSVS+VPPAY
Sbjct: 873 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADGIQSLTNNLCYTYARCTRSVSIVPPAY 932
Query: 181 YAHLAAFRARFYMEPEMSENQTSKSSNGTNGGL-----VKPLPAVKEKVKRVMFYC 333
YAHLAAFRARFY+EPE+ ++ S T VKPLPA+KE VKRVMFYC
Sbjct: 933 YAHLAAFRARFYLEPEIMQDNGSPGKKNTKTTTVGDVGVKPLPALKENVKRVMFYC 988
>sptr|Q8LNZ9|Q8LNZ9 AGO1 homologous protein.
Length = 909
Score = 186 bits (472), Expect = 2e-46
Identities = 89/126 (70%), Positives = 99/126 (78%), Gaps = 15/126 (11%)
Frame = +1
Query: 1 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAY 180
EFDFYLCSHAGI+GTSRPAHYHVLW+ENNFTAD +Q LTNNLCYTYARCTRSVS+VPPAY
Sbjct: 784 EFDFYLCSHAGIKGTSRPAHYHVLWNENNFTADALQILTNNLCYTYARCTRSVSIVPPAY 843
Query: 181 YAHLAAFRARFYMEPEMSENQTSKSSNGTNGGL---------------VKPLPAVKEKVK 315
YAHLAAFRARFYMEP+ S++ + S G G L VKPLPA+K+ VK
Sbjct: 844 YAHLAAFRARFYMEPDTSDSSSVVSGPGVRGPLSGSSTSRTRAPGGAAVKPLPALKDSVK 903
Query: 316 RVMFYC 333
RVMFYC
Sbjct: 904 RVMFYC 909
>sw|Q9SJK3|AGOL_ARATH Argonaute-like protein At2g27880.
Length = 997
Score = 177 bits (449), Expect = 1e-43
Identities = 83/113 (73%), Positives = 96/113 (84%), Gaps = 2/113 (1%)
Frame = +1
Query: 1 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAY 180
EFDFYL SHAGIQGTSRPAHYHVL DEN FTAD++Q LTNNLCYTYARCT+SVS+VPPAY
Sbjct: 885 EFDFYLNSHAGIQGTSRPAHYHVLLDENGFTADQLQMLTNNLCYTYARCTKSVSIVPPAY 944
Query: 181 YAHLAAFRARFYMEPEMSENQTSKSSNGTN--GGLVKPLPAVKEKVKRVMFYC 333
YAHLAAFRAR+YME EMS+ +S+S + T G ++ LPA+K+ VK VMFYC
Sbjct: 945 YAHLAAFRARYYMESEMSDGGSSRSRSSTTGVGQVISQLPAIKDNVKEVMFYC 997
>sptr|Q7XSA2|Q7XSA2 OSJNBa0005N02.3 protein.
Length = 1254
Score = 171 bits (432), Expect = 9e-42
Identities = 78/93 (83%), Positives = 84/93 (90%)
Frame = +1
Query: 1 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAY 180
EFDFYLCSHAGIQGTSRPAHYHVLWDEN FTADE+QTLTNNLCYTYARCTRSVS+VPPAY
Sbjct: 1080 EFDFYLCSHAGIQGTSRPAHYHVLWDENKFTADELQTLTNNLCYTYARCTRSVSIVPPAY 1139
Query: 181 YAHLAAFRARFYMEPEMSENQTSKSSNGTNGGL 279
YAHLAAFRARFYMEPE S++ + S T+ GL
Sbjct: 1140 YAHLAAFRARFYMEPETSDSGSMASGAATSRGL 1172
>sptr|Q851R2|Q851R2 Putative argonaute protein.
Length = 1058
Score = 170 bits (431), Expect = 1e-41
Identities = 82/116 (70%), Positives = 96/116 (82%), Gaps = 5/116 (4%)
Frame = +1
Query: 1 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAY 180
EFDFYLCSHAGIQGTSRP HYHVL+DEN+FTAD +Q+LTNNLCYTYARCTR+VSVVPPAY
Sbjct: 943 EFDFYLCSHAGIQGTSRPTHYHVLYDENHFTADALQSLTNNLCYTYARCTRAVSVVPPAY 1002
Query: 181 YAHLAAFRARFYMEPEMSENQTSKSSNG----TNGGL-VKPLPAVKEKVKRVMFYC 333
YAHLAAFRAR+Y+E E S+ ++ S+G G + V+ LP +KE VK VMFYC
Sbjct: 1003 YAHLAAFRARYYVEGESSDGGSTPGSSGQAVAREGPVEVRQLPKIKENVKDVMFYC 1058
>sptrnew|BAC83909|BAC83909 Putative leaf development protein
Argonaute.
Length = 1052
Score = 166 bits (419), Expect = 3e-40
Identities = 82/117 (70%), Positives = 90/117 (76%), Gaps = 6/117 (5%)
Frame = +1
Query: 1 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAY 180
EFDFYLCSHAGIQGTSRP HYHVL DEN F+AD++Q LT NLCYTYARCTRSVSVVPPAY
Sbjct: 936 EFDFYLCSHAGIQGTSRPTHYHVLHDENRFSADQLQMLTYNLCYTYARCTRSVSVVPPAY 995
Query: 181 YAHLAAFRARFYMEPEMSENQTSKSSNGTN---GG---LVKPLPAVKEKVKRVMFYC 333
YAHLAAFRAR+Y EP + +S S G GG V+ LP +KE VK VMFYC
Sbjct: 996 YAHLAAFRARYYDEPPAMDGASSVGSGGNQAAAGGQPPAVRRLPQIKENVKDVMFYC 1052
>sptr|Q7Y001|Q7Y001 Putative leaf development and shoot apical
meristem regulating protein.
Length = 1055
Score = 165 bits (417), Expect = 5e-40
Identities = 77/111 (69%), Positives = 86/111 (77%)
Frame = +1
Query: 1 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAY 180
EFDFYLCSH+GI+GTSRP HYHVL DEN F AD +QTLT NL YTYARCTR+VS+VPPAY
Sbjct: 945 EFDFYLCSHSGIKGTSRPTHYHVLLDENGFKADTLQTLTYNLSYTYARCTRAVSIVPPAY 1004
Query: 181 YAHLAAFRARFYMEPEMSENQTSKSSNGTNGGLVKPLPAVKEKVKRVMFYC 333
YAHL AFRAR+YME E S+ +S S KPLP +KE VKR MFYC
Sbjct: 1005 YAHLGAFRARYYMEDEHSDQGSSSSVTTRTDRSTKPLPEIKENVKRFMFYC 1055
>sptr|Q7XZZ9|Q7XZZ9 Putative leaf development and shoot apical
meristem regulating protein.
Length = 895
Score = 157 bits (397), Expect = 1e-37
Identities = 75/118 (63%), Positives = 88/118 (74%), Gaps = 7/118 (5%)
Frame = +1
Query: 1 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAY 180
EFDFYLCSH+GIQGTS P HY+VL+DENNF+AD +QTLT +LCYTYARCTRSVS+VPP Y
Sbjct: 778 EFDFYLCSHSGIQGTSHPTHYYVLFDENNFSADALQTLTYHLCYTYARCTRSVSIVPPVY 837
Query: 181 YAHLAAFRARFYMEPEMSENQTSKSSNGTNGGL-------VKPLPAVKEKVKRVMFYC 333
YAHLAA RAR Y+E + S S++ G VKPLP +KE VK+ MFYC
Sbjct: 838 YAHLAASRARHYLEEGSLPDHGSSSASAAGGSRRNDRGVPVKPLPEIKENVKQFMFYC 895
>sptrnew|AAQ92355|AAQ92355 ZIPPY.
Length = 990
Score = 147 bits (371), Expect = 1e-34
Identities = 67/114 (58%), Positives = 84/114 (73%), Gaps = 3/114 (2%)
Frame = +1
Query: 1 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAY 180
EFDFYLCSH G++GTSRP HYH+LWDEN FT+DE+Q L NLCYT+ RCT+ +S+VPPAY
Sbjct: 877 EFDFYLCSHLGVKGTSRPTHYHILWDENEFTSDELQRLVYNLCYTFVRCTKPISIVPPAY 936
Query: 181 YAHLAAFRARFYMEPEMSENQTSKS-SNGTNGGLVK--PLPAVKEKVKRVMFYC 333
YAHLAA+R R Y+E N S + S+ + G K PLP + + VK +MFYC
Sbjct: 937 YAHLAAYRGRLYIERSSESNGGSMNPSSVSRVGPPKTIPLPKLSDNVKNLMFYC 990
>sptr|Q9C793|Q9C793 Pinhead-like protein.
Length = 990
Score = 147 bits (371), Expect = 1e-34
Identities = 67/114 (58%), Positives = 84/114 (73%), Gaps = 3/114 (2%)
Frame = +1
Query: 1 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAY 180
EFDFYLCSH G++GTSRP HYH+LWDEN FT+DE+Q L NLCYT+ RCT+ +S+VPPAY
Sbjct: 877 EFDFYLCSHLGVKGTSRPTHYHILWDENEFTSDELQRLVYNLCYTFVRCTKPISIVPPAY 936
Query: 181 YAHLAAFRARFYMEPEMSENQTSKS-SNGTNGGLVK--PLPAVKEKVKRVMFYC 333
YAHLAA+R R Y+E N S + S+ + G K PLP + + VK +MFYC
Sbjct: 937 YAHLAAYRGRLYIERSSESNGGSMNPSSVSRVGPPKTIPLPKLSDNVKNLMFYC 990
>sw|O77503|I2C2_RABIT Eukaryotic translation initiation factor 2C 2
(eIF2C 2) (eIF-2C 2).
Length = 813
Score = 135 bits (341), Expect = 3e-31
Identities = 65/114 (57%), Positives = 83/114 (72%), Gaps = 4/114 (3%)
Frame = +1
Query: 1 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAY 180
EFDFYLCSHAGIQGTSRP+HYHVLWD+N F++DE+Q LT LC+TY RCTRSVS+ PAY
Sbjct: 699 EFDFYLCSHAGIQGTSRPSHYHVLWDDNRFSSDELQILTYQLCHTYVRCTRSVSIPAPAY 758
Query: 181 YAHLAAFRARFYM---EPEMSE-NQTSKSSNGTNGGLVKPLPAVKEKVKRVMFY 330
YAHL AFRAR+++ E + +E + TS SNG + + V + R M++
Sbjct: 759 YAHLVAFRARYHLVDKEHDSAEGSHTSGQSNGRDHQALAKAVQVHQDTLRTMYF 812
>sptr|Q8WV58|Q8WV58 Hypothetical protein (Eukaryotic translation
initiation factor 2C, 2).
Length = 585
Score = 135 bits (341), Expect = 3e-31
Identities = 65/114 (57%), Positives = 83/114 (72%), Gaps = 4/114 (3%)
Frame = +1
Query: 1 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAY 180
EFDFYLCSHAGIQGTSRP+HYHVLWD+N F++DE+Q LT LC+TY RCTRSVS+ PAY
Sbjct: 471 EFDFYLCSHAGIQGTSRPSHYHVLWDDNRFSSDELQILTYQLCHTYVRCTRSVSIPAPAY 530
Query: 181 YAHLAAFRARFYM---EPEMSE-NQTSKSSNGTNGGLVKPLPAVKEKVKRVMFY 330
YAHL AFRAR+++ E + +E + TS SNG + + V + R M++
Sbjct: 531 YAHLVAFRARYHLVDKEHDSAEGSHTSGQSNGRDHQALAKAVQVHQDTLRTMYF 584
>sw|Q9QZ81|I2C2_RAT Eukaryotic translation initiation factor 2C 2
(eIF2C 2) (eIF-2C 2) (Golgi ER protein 95 kDa) (GERp95).
Length = 863
Score = 135 bits (341), Expect = 3e-31
Identities = 65/114 (57%), Positives = 83/114 (72%), Gaps = 4/114 (3%)
Frame = +1
Query: 1 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAY 180
EFDFYLCSHAGIQGTSRP+HYHVLWD+N F++DE+Q LT LC+TY RCTRSVS+ PAY
Sbjct: 749 EFDFYLCSHAGIQGTSRPSHYHVLWDDNRFSSDELQILTYQLCHTYVRCTRSVSIPAPAY 808
Query: 181 YAHLAAFRARFYM---EPEMSE-NQTSKSSNGTNGGLVKPLPAVKEKVKRVMFY 330
YAHL AFRAR+++ E + +E + TS SNG + + V + R M++
Sbjct: 809 YAHLVAFRARYHLVDKEHDSAEGSHTSGQSNGRDHQALAKAVQVHQDTLRTMYF 862
>sptrnew|BAC98205|BAC98205 MKIAA1567 protein (Fragment).
Length = 703
Score = 135 bits (341), Expect = 3e-31
Identities = 65/114 (57%), Positives = 83/114 (72%), Gaps = 4/114 (3%)
Frame = +1
Query: 1 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAY 180
EFDFYLCSHAGIQGTSRP+HYHVLWD+N F++DE+Q LT LC+TY RCTRSVS+ PAY
Sbjct: 589 EFDFYLCSHAGIQGTSRPSHYHVLWDDNRFSSDELQILTYQLCHTYVRCTRSVSIPAPAY 648
Query: 181 YAHLAAFRARFYM---EPEMSE-NQTSKSSNGTNGGLVKPLPAVKEKVKRVMFY 330
YAHL AFRAR+++ E + +E + TS SNG + + V + R M++
Sbjct: 649 YAHLVAFRARYHLVDKEHDSAEGSHTSGQSNGRDHQALAKAVQVHQDTLRTMYF 702
>sptr|Q96ID1|Q96ID1 Hypothetical protein.
Length = 377
Score = 135 bits (341), Expect = 3e-31
Identities = 65/114 (57%), Positives = 83/114 (72%), Gaps = 4/114 (3%)
Frame = +1
Query: 1 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAY 180
EFDFYLCSHAGIQGTSRP+HYHVLWD+N F++DE+Q LT LC+TY RCTRSVS+ PAY
Sbjct: 263 EFDFYLCSHAGIQGTSRPSHYHVLWDDNRFSSDELQILTYQLCHTYVRCTRSVSIPAPAY 322
Query: 181 YAHLAAFRARFYM---EPEMSE-NQTSKSSNGTNGGLVKPLPAVKEKVKRVMFY 330
YAHL AFRAR+++ E + +E + TS SNG + + V + R M++
Sbjct: 323 YAHLVAFRARYHLVDKEHDSAEGSHTSGQSNGRDHQALAKAVQVHQDTLRTMYF 376
>sw|Q9UKV8|I2C2_HUMAN Eukaryotic translation initiation factor 2C 2
(eIF2C 2) (eIF-2C 2) (Fragment).
Length = 377
Score = 135 bits (341), Expect = 3e-31
Identities = 65/114 (57%), Positives = 83/114 (72%), Gaps = 4/114 (3%)
Frame = +1
Query: 1 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAY 180
EFDFYLCSHAGIQGTSRP+HYHVLWD+N F++DE+Q LT LC+TY RCTRSVS+ PAY
Sbjct: 263 EFDFYLCSHAGIQGTSRPSHYHVLWDDNRFSSDELQILTYQLCHTYVRCTRSVSIPAPAY 322
Query: 181 YAHLAAFRARFYM---EPEMSE-NQTSKSSNGTNGGLVKPLPAVKEKVKRVMFY 330
YAHL AFRAR+++ E + +E + TS SNG + + V + R M++
Sbjct: 323 YAHLVAFRARYHLVDKEHDSAEGSHTSGQSNGRDHQALAKAVQVHQDTLRTMYF 376
>sptr|Q8TCZ5|Q8TCZ5 Eukaryotic initiation factor 2C2.
Length = 851
Score = 135 bits (341), Expect = 3e-31
Identities = 65/114 (57%), Positives = 83/114 (72%), Gaps = 4/114 (3%)
Frame = +1
Query: 1 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAY 180
EFDFYLCSHAGIQGTSRP+HYHVLWD+N F++DE+Q LT LC+TY RCTRSVS+ PAY
Sbjct: 737 EFDFYLCSHAGIQGTSRPSHYHVLWDDNRFSSDELQILTYQLCHTYVRCTRSVSIPAPAY 796
Query: 181 YAHLAAFRARFYM---EPEMSE-NQTSKSSNGTNGGLVKPLPAVKEKVKRVMFY 330
YAHL AFRAR+++ E + +E + TS SNG + + V + R M++
Sbjct: 797 YAHLVAFRARYHLVDKEHDSAEGSHTSGQSNGRDHQALAKAVQVHQDTLRTMYF 850
>sptrnew|AAH56639|AAH56639 Hypothetical protein (Fragment).
Length = 437
Score = 135 bits (341), Expect = 3e-31
Identities = 65/114 (57%), Positives = 83/114 (72%), Gaps = 4/114 (3%)
Frame = +1
Query: 1 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAY 180
EFDFYLCSHAGIQGTSRP+HYHVLWD+N F++DE+Q LT LC+TY RCTRSVS+ PAY
Sbjct: 323 EFDFYLCSHAGIQGTSRPSHYHVLWDDNRFSSDELQILTYQLCHTYVRCTRSVSIPAPAY 382
Query: 181 YAHLAAFRARFYM---EPEMSE-NQTSKSSNGTNGGLVKPLPAVKEKVKRVMFY 330
YAHL AFRAR+++ E + +E + TS SNG + + V + R M++
Sbjct: 383 YAHLVAFRARYHLVDKEHDSAEGSHTSGQSNGRDHQALAKAVQVHQDTLRTMYF 436
>sptr|Q8R3Q7|Q8R3Q7 Similar to eukaryotic translation initiation
factor 2C, 2 (Fragment).
Length = 530
Score = 135 bits (341), Expect = 3e-31
Identities = 65/114 (57%), Positives = 83/114 (72%), Gaps = 4/114 (3%)
Frame = +1
Query: 1 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAY 180
EFDFYLCSHAGIQGTSRP+HYHVLWD+N F++DE+Q LT LC+TY RCTRSVS+ PAY
Sbjct: 416 EFDFYLCSHAGIQGTSRPSHYHVLWDDNRFSSDELQILTYQLCHTYVRCTRSVSIPAPAY 475
Query: 181 YAHLAAFRARFYM---EPEMSE-NQTSKSSNGTNGGLVKPLPAVKEKVKRVMFY 330
YAHL AFRAR+++ E + +E + TS SNG + + V + R M++
Sbjct: 476 YAHLVAFRARYHLVDKEHDSAEGSHTSGQSNGRDHQALAKAVQVHQDTLRTMYF 529
>sptr|Q8CJF9|Q8CJF9 Piwi/Argonaute family protain meIF2C3.
Length = 860
Score = 135 bits (339), Expect = 5e-31
Identities = 65/114 (57%), Positives = 82/114 (71%), Gaps = 4/114 (3%)
Frame = +1
Query: 1 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAY 180
EFDFYLCSHAGIQGTSRP+HYHVLWD+N FTADE+Q LT LC+TY RCTRSVS+ PAY
Sbjct: 746 EFDFYLCSHAGIQGTSRPSHYHVLWDDNFFTADELQLLTYQLCHTYVRCTRSVSIPAPAY 805
Query: 181 YAHLAAFRARFYM---EPEMSE-NQTSKSSNGTNGGLVKPLPAVKEKVKRVMFY 330
YAHL AFRAR+++ E + +E + S SNG + + + + R M++
Sbjct: 806 YAHLVAFRARYHLVDKEHDSAEGSHVSGQSNGRDPQALAKAVQIHQDTLRTMYF 859
>sptr|Q9H9G7|Q9H9G7 Hypothetical protein FLJ12765.
Length = 860
Score = 135 bits (339), Expect = 5e-31
Identities = 65/114 (57%), Positives = 82/114 (71%), Gaps = 4/114 (3%)
Frame = +1
Query: 1 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAY 180
EFDFYLCSHAGIQGTSRP+HYHVLWD+N FTADE+Q LT LC+TY RCTRSVS+ PAY
Sbjct: 746 EFDFYLCSHAGIQGTSRPSHYHVLWDDNCFTADELQLLTYQLCHTYVRCTRSVSIPAPAY 805
Query: 181 YAHLAAFRARFYM---EPEMSE-NQTSKSSNGTNGGLVKPLPAVKEKVKRVMFY 330
YAHL AFRAR+++ E + +E + S SNG + + + + R M++
Sbjct: 806 YAHLVAFRARYHLVDKEHDSAEGSHVSGQSNGRDPQALAKAVQIHQDTLRTMYF 859
>sptr|Q9H1U6|Q9H1U6 DJ665N4.6 (Novel protein) (Fragment).
Length = 640
Score = 135 bits (339), Expect = 5e-31
Identities = 65/114 (57%), Positives = 82/114 (71%), Gaps = 4/114 (3%)
Frame = +1
Query: 1 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAY 180
EFDFYLCSHAGIQGTSRP+HYHVLWD+N FTADE+Q LT LC+TY RCTRSVS+ PAY
Sbjct: 526 EFDFYLCSHAGIQGTSRPSHYHVLWDDNCFTADELQLLTYQLCHTYVRCTRSVSIPAPAY 585
Query: 181 YAHLAAFRARFYM---EPEMSE-NQTSKSSNGTNGGLVKPLPAVKEKVKRVMFY 330
YAHL AFRAR+++ E + +E + S SNG + + + + R M++
Sbjct: 586 YAHLVAFRARYHLVDKEHDSAEGSHVSGQSNGRDPQALAKAVQIHQDTLRTMYF 639
>sptrnew|DAA00372|DAA00372 Argonaute 3.
Length = 748
Score = 135 bits (339), Expect = 5e-31
Identities = 65/114 (57%), Positives = 82/114 (71%), Gaps = 4/114 (3%)
Frame = +1
Query: 1 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAY 180
EFDFYLCSHAGIQGTSRP+HYHVLWD+N FTADE+Q LT LC+TY RCTRSVS+ PAY
Sbjct: 634 EFDFYLCSHAGIQGTSRPSHYHVLWDDNFFTADELQLLTYQLCHTYVRCTRSVSIPAPAY 693
Query: 181 YAHLAAFRARFYM---EPEMSE-NQTSKSSNGTNGGLVKPLPAVKEKVKRVMFY 330
YAHL AFRAR+++ E + +E + S SNG + + + + R M++
Sbjct: 694 YAHLVAFRARYHLVDKEHDSAEGSHVSGQSNGRDPQALAKAVQIHQDTLRTMYF 747
>sptr|Q8CJG0|Q8CJG0 Piwi/Argonaute family protain meIF2C2.
Length = 860
Score = 134 bits (337), Expect = 9e-31
Identities = 64/114 (56%), Positives = 82/114 (71%), Gaps = 4/114 (3%)
Frame = +1
Query: 1 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAY 180
EFDFYLCSHAGIQGT RP+HYHVLWD+N F++DE+Q LT LC+TY RCTRSVS+ PAY
Sbjct: 746 EFDFYLCSHAGIQGTGRPSHYHVLWDDNRFSSDELQILTYQLCHTYVRCTRSVSIPAPAY 805
Query: 181 YAHLAAFRARFYM---EPEMSE-NQTSKSSNGTNGGLVKPLPAVKEKVKRVMFY 330
YAHL AFRAR+++ E + +E + TS SNG + + V + R M++
Sbjct: 806 YAHLVAFRARYHLVDKEHDSAEGSHTSGQSNGRDHQALAKAVQVHQDTLRTMYF 859
>sptr|Q20578|Q20578 F48F7.1 protein.
Length = 1002
Score = 134 bits (336), Expect = 1e-30
Identities = 58/90 (64%), Positives = 71/90 (78%)
Frame = +1
Query: 1 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAY 180
EFDFYLCSHAGIQGTSRP+HYHVLWD+NN TADE+Q LT +C+TY RCTRSVS+ PAY
Sbjct: 888 EFDFYLCSHAGIQGTSRPSHYHVLWDDNNLTADELQQLTYQMCHTYVRCTRSVSIPAPAY 947
Query: 181 YAHLAAFRARFYMEPEMSENQTSKSSNGTN 270
YAHL AFRAR+++ ++ +GT+
Sbjct: 948 YAHLVAFRARYHLVDREHDSGEGSQPSGTS 977
>sptr|Q9V3L2|Q9V3L2 AGO1 protein.
Length = 950
Score = 132 bits (333), Expect = 3e-30
Identities = 64/115 (55%), Positives = 84/115 (73%), Gaps = 5/115 (4%)
Frame = +1
Query: 1 EFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAY 180
EFDFYLCSH GIQGTSRP+HYHVLWD+N+F +DE+Q LT LC+TY RCTRSVS+ PAY
Sbjct: 836 EFDFYLCSHQGIQGTSRPSHYHVLWDDNHFDSDELQCLTYQLCHTYVRCTRSVSIPAPAY 895
Query: 181 YAHLAAFRARFYM---EPEMSE--NQTSKSSNGTNGGLVKPLPAVKEKVKRVMFY 330
YAHL AFRAR+++ E + E +Q+ S + T G + + + V K+VM++
Sbjct: 896 YAHLVAFRARYHLVEKEHDSGEGSHQSGCSEDRTPGAMARAI-TVHADTKKVMYF 949
Database: /db/trembl-ebi/tmp/swall
Posted date: Oct 24, 2003 8:22 PM
Number of letters in database: 411,996,502
Number of sequences in database: 1,288,562
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 490,450,884
Number of Sequences: 1288562
Number of extensions: 10108278
Number of successful extensions: 23823
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 23062
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 23797
length of database: 411,996,502
effective HSP length: 117
effective length of database: 261,234,748
effective search space used: 24556066312
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

