BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
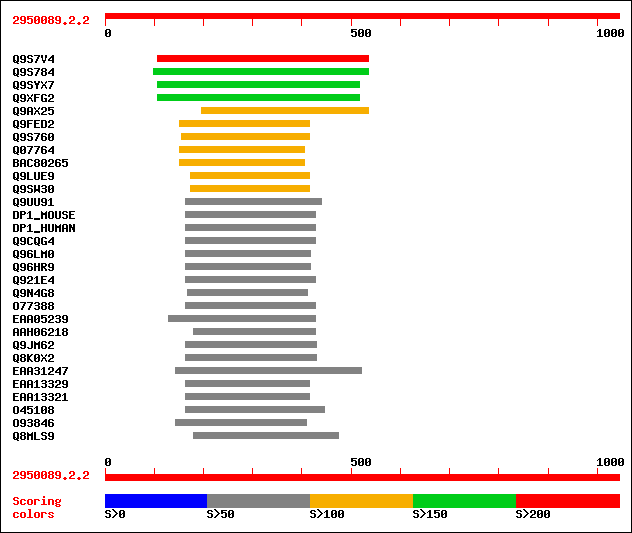
Query= 2950089.2.2
(1044 letters)
Database: /db/trembl-ebi/tmp/swall
1,272,877 sequences; 406,886,216 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sptr|Q9S7V4|Q9S7V4 ATHVA22A. 220 2e-56
sptr|Q9S784|Q9S784 ATHVA22C. 180 3e-44
sptr|Q9SYX7|Q9SYX7 AtHVA22b. 165 1e-39
sptr|Q9XFG2|Q9XFG2 ATHVA22B. 164 1e-39
sptr|Q9AX25|Q9AX25 P0456A01.10 protein (P0042A10.34 protein). 122 1e-26
sptr|Q9FED2|Q9FED2 AtHVA22e (Abscisic acid-induced-like protein). 117 2e-25
sptr|Q9S760|Q9S760 ATHVA22D (Putative abscisic acid-induced prot... 112 9e-24
sptr|Q07764|Q07764 Abscisic acid and stress inducible (A22) gene. 112 1e-23
sptrnew|BAC80265|BAC80265 Hypothetical protein Wrab15. 111 1e-23
sptr|Q9LUE9|Q9LUE9 Genomic DNA, chromosome 5, P1 clone:MFB16. 108 1e-22
sptr|Q9SW30|Q9SW30 Abscisic acid-induced-like protein. 104 2e-21
sptr|Q9UU91|Q9UU91 Putative transport protein. 79 1e-13
sw|Q60870|DP1_MOUSE Polyposis locus protein 1 homolog (TB2 prote... 77 5e-13
sw|Q00765|DP1_HUMAN Polyposis locus protein 1 (TB2 protein). 76 7e-13
sptr|Q9CQG4|Q9CQG4 Deleted in polyposis 1. 75 2e-12
sptr|Q96LM0|Q96LM0 Hypothetical protein FLJ25383 (TB2 protein-li... 74 3e-12
sptr|Q96HR9|Q96HR9 Hypothetical protein. 74 3e-12
sptr|Q921E4|Q921E4 Deleted in polyposis 1. 74 4e-12
sptr|Q9N4G8|Q9N4G8 Hypothetical 20.6 kDa protein. 73 6e-12
sptr|O77388|O77388 Hypothetical 26.0 kDa protein. 73 6e-12
sptrnew|EAA05239|EAA05239 ENSANGP00000018512 (Fragment). 73 8e-12
sptrnew|AAH06218|AAH06218 Hypothetical protein. 72 1e-11
sptr|Q9JM62|Q9JM62 Polyposis locus protein 1-like 1 (TB2 protein... 72 2e-11
sptr|Q8K0X2|Q8K0X2 Hypothetical protein. 72 2e-11
sptrnew|EAA31247|EAA31247 Hypothetical protein. 71 2e-11
sptrnew|EAA13329|EAA13329 ENSANGP00000003353 (Fragment). 71 3e-11
sptrnew|EAA13321|EAA13321 AgCP2771 (Fragment). 71 3e-11
sptr|O45108|O45108 Hypothetical 22.8 kDa protein. 69 1e-10
sptr|O93846|O93846 Pathogenicity protein. 69 1e-10
sptr|Q8MLS9|Q8MLS9 CG30193-PA (RE29641p). 68 2e-10
>sptr|Q9S7V4|Q9S7V4 ATHVA22A.
Length = 177
Score = 220 bits (561), Expect = 2e-56
Identities = 98/144 (68%), Positives = 123/144 (85%), Gaps = 1/144 (0%)
Frame = +2
Query: 107 GSGSFLKVLAKNFDVLAGPLISLAYPLYASVRAIETKNPVDDQQWLTYWVLYSFITLFEL 286
G+G+FLKVL +NFDVLAGP++SL YPLYASV+AIET++ DD+QWLTYWVLYS +TL EL
Sbjct: 4 GAGNFLKVLLRNFDVLAGPVVSLVYPLYASVQAIETQSHADDKQWLTYWVLYSLLTLIEL 63
Query: 287 TFAPIIEWLPFWSYAKLFFNCWLVLPWFNGAAYVYDHFVRPMFVNRQIVNIWYIPRN-EK 463
TFA +IEWLP WSY KL CWLV+P+F+GAAYVY+HFVRP+FVN + +NIWY+P+ +
Sbjct: 64 TFAKLIEWLPIWSYMKLILTCWLVIPYFSGAAYVYEHFVRPVFVNPRSINIWYVPKKMDI 123
Query: 464 LGKSDDVLSAAERYIEQNGPEAFE 535
K DDVL+AAE+YI +NGP+AFE
Sbjct: 124 FRKPDDVLTAAEKYIAENGPDAFE 147
>sptr|Q9S784|Q9S784 ATHVA22C.
Length = 184
Score = 180 bits (456), Expect = 3e-44
Identities = 85/152 (55%), Positives = 109/152 (71%), Gaps = 6/152 (3%)
Frame = +2
Query: 98 SKMGSGSFLKVLAKNFDVLAGPLISLAYPLYASVRAIETKNPVDDQQWLTYWVLYSFITL 277
S G + L+VL KNFDVLA PL++L YPLYASV+AIET++ +D+QWLTYWVLY+ I+L
Sbjct: 3 SNSGDDNVLQVLIKNFDVLALPLVTLVYPLYASVKAIETRSLPEDEQWLTYWVLYALISL 62
Query: 278 FELTFAPIIEWLPFWSYAKLFFNCWLVLPWFNGAAYVYDHFVRPMF--VNRQIVNIWYIP 451
FELTF+ +EW P W Y KLF CWLVLP FNGA ++Y HF+RP + R IWY+P
Sbjct: 63 FELTFSKPLEWFPIWPYMKLFGICWLVLPQFNGAEHIYKHFIRPFYRDPQRATTKIWYVP 122
Query: 452 RNE----KLGKSDDVLSAAERYIEQNGPEAFE 535
+ DD+L+AAE+Y+EQ+G EAFE
Sbjct: 123 HKKFNFFPKRDDDDILTAAEKYMEQHGTEAFE 154
>sptr|Q9SYX7|Q9SYX7 AtHVA22b.
Length = 167
Score = 165 bits (417), Expect = 1e-39
Identities = 76/137 (55%), Positives = 99/137 (72%)
Frame = +2
Query: 107 GSGSFLKVLAKNFDVLAGPLISLAYPLYASVRAIETKNPVDDQQWLTYWVLYSFITLFEL 286
G GS +KV+ KNFDV+AGP+ISL YPLYASVRAIE+++ DD+QWLTYW LYS I LFEL
Sbjct: 4 GIGSLVKVIFKNFDVIAGPMISLVYPLYASVRAIESRSHGDDKQWLTYWALYSLIKLFEL 63
Query: 287 TFAPIIEWLPFWSYAKLFFNCWLVLPWFNGAAYVYDHFVRPMFVNRQIVNIWYIPRNEKL 466
TF ++EW+P + YAKL WLVLP NGAAY+Y+H+VR ++ VN+WY+P
Sbjct: 64 TFFRLLEWIPLYPYAKLALTSWLVLPGMNGAAYLYEHYVRSFLLSPHTVNVWYVPAK--- 120
Query: 467 GKSDDVLSAAERYIEQN 517
K DD+ + A ++ N
Sbjct: 121 -KDDDLGATAGKFTPVN 136
>sptr|Q9XFG2|Q9XFG2 ATHVA22B.
Length = 167
Score = 164 bits (416), Expect = 1e-39
Identities = 76/137 (55%), Positives = 99/137 (72%)
Frame = +2
Query: 107 GSGSFLKVLAKNFDVLAGPLISLAYPLYASVRAIETKNPVDDQQWLTYWVLYSFITLFEL 286
G GS +KV+ KNFDV+AGP+ISL YPLYASVRAIE+++ DD+QWLTYW LYS I LFEL
Sbjct: 4 GIGSLVKVIFKNFDVIAGPVISLVYPLYASVRAIESRSHGDDKQWLTYWALYSLIKLFEL 63
Query: 287 TFAPIIEWLPFWSYAKLFFNCWLVLPWFNGAAYVYDHFVRPMFVNRQIVNIWYIPRNEKL 466
TF ++EW+P + YAKL WLVLP NGAAY+Y+H+VR ++ VN+WY+P
Sbjct: 64 TFFRLLEWIPLYPYAKLALTSWLVLPGMNGAAYLYEHYVRSFLLSPHTVNVWYVPAK--- 120
Query: 467 GKSDDVLSAAERYIEQN 517
K DD+ + A ++ N
Sbjct: 121 -KDDDLGATAGKFTPVN 136
>sptr|Q9AX25|Q9AX25 P0456A01.10 protein (P0042A10.34 protein).
Length = 128
Score = 122 bits (305), Expect = 1e-26
Identities = 60/115 (52%), Positives = 77/115 (66%), Gaps = 2/115 (1%)
Frame = +2
Query: 197 VRAIETKNPVDDQQWLTYWVLYSFITLFELTFAPIIEWLPFWSYAKLFFNCWLVLPWFNG 376
+RAIE+ + +DDQQWLTYWVLYS ITLFEL+ +++W P W Y KL F CWLVLP FNG
Sbjct: 1 MRAIESPSTLDDQQWLTYWVLYSLITLFELSCWKVLQWFPLWPYMKLLFCCWLVLPIFNG 60
Query: 377 AAYVYDHFVRPMFVNRQIVNIWYIPRNEKLGK--SDDVLSAAERYIEQNGPEAFE 535
AAY+Y+ VR F Q V+ Y R K + S D + ER+IE +GP+A +
Sbjct: 61 AAYIYETHVRRYFKIGQYVSPNYNERQRKALQMMSLDARKSVERFIESHGPDALD 115
>sptr|Q9FED2|Q9FED2 AtHVA22e (Abscisic acid-induced-like protein).
Length = 116
Score = 117 bits (294), Expect = 2e-25
Identities = 52/88 (59%), Positives = 65/88 (73%)
Frame = +2
Query: 152 LAGPLISLAYPLYASVRAIETKNPVDDQQWLTYWVLYSFITLFELTFAPIIEWLPFWSYA 331
LAGP++ L YPLYASV AIE+ + VDD+QWL YW+LYSF+TL EL ++EW+P W A
Sbjct: 14 LAGPVVMLLYPLYASVIAIESPSKVDDEQWLAYWILYSFLTLSELILQSLLEWIPIWYTA 73
Query: 332 KLFFNCWLVLPWFNGAAYVYDHFVRPMF 415
KL F WLVLP F GAA++Y+ VR F
Sbjct: 74 KLVFVAWLVLPQFRGAAFIYNKVVREQF 101
>sptr|Q9S760|Q9S760 ATHVA22D (Putative abscisic acid-induced
protein).
Length = 135
Score = 112 bits (280), Expect = 9e-24
Identities = 48/87 (55%), Positives = 62/87 (71%)
Frame = +2
Query: 155 AGPLISLAYPLYASVRAIETKNPVDDQQWLTYWVLYSFITLFELTFAPIIEWLPFWSYAK 334
AGP++ L YPLYASV A+E+ VDD+QWL YW++YSF++L EL +IEW+P W K
Sbjct: 15 AGPIVMLLYPLYASVIAMESTTKVDDEQWLAYWIIYSFLSLTELILQSLIEWIPIWYTVK 74
Query: 335 LFFNCWLVLPWFNGAAYVYDHFVRPMF 415
L F WLVLP F GAA++Y+ VR F
Sbjct: 75 LVFVAWLVLPQFQGAAFIYNRVVREQF 101
>sptr|Q07764|Q07764 Abscisic acid and stress inducible (A22) gene.
Length = 130
Score = 112 bits (279), Expect = 1e-23
Identities = 48/85 (56%), Positives = 62/85 (72%)
Frame = +2
Query: 152 LAGPLISLAYPLYASVRAIETKNPVDDQQWLTYWVLYSFITLFELTFAPIIEWLPFWSYA 331
+AGP I+L YPLYASV A+E+ + VDD+QWL YW+LYSFITL E+ P++ W+P W
Sbjct: 14 VAGPSITLLYPLYASVCAMESPSKVDDEQWLAYWILYSFITLLEMVAEPVLYWIPVWYPV 73
Query: 332 KLFFNCWLVLPWFNGAAYVYDHFVR 406
KL F WL LP F GA+++YD VR
Sbjct: 74 KLLFVAWLALPQFKGASFIYDKVVR 98
>sptrnew|BAC80265|BAC80265 Hypothetical protein Wrab15.
Length = 130
Score = 111 bits (278), Expect = 1e-23
Identities = 48/85 (56%), Positives = 62/85 (72%)
Frame = +2
Query: 152 LAGPLISLAYPLYASVRAIETKNPVDDQQWLTYWVLYSFITLFELTFAPIIEWLPFWSYA 331
+AGP I+L YPLYASV A+E+ + VDD+QWL YW+LYSFITL E+ P++ W+P W
Sbjct: 14 VAGPSITLLYPLYASVCAMESPSKVDDEQWLAYWILYSFITLMEMLAEPVLYWIPVWYPV 73
Query: 332 KLFFNCWLVLPWFNGAAYVYDHFVR 406
KL F WL LP F GA+++YD VR
Sbjct: 74 KLLFVAWLALPQFKGASFIYDKVVR 98
>sptr|Q9LUE9|Q9LUE9 Genomic DNA, chromosome 5, P1 clone:MFB16.
Length = 107
Score = 108 bits (270), Expect = 1e-22
Identities = 48/81 (59%), Positives = 59/81 (72%)
Frame = +2
Query: 173 LAYPLYASVRAIETKNPVDDQQWLTYWVLYSFITLFELTFAPIIEWLPFWSYAKLFFNCW 352
L YPLYASV AIE+ + VDD+QWL YW+LYSF+TL EL ++EW+P W AKL F W
Sbjct: 2 LLYPLYASVIAIESPSKVDDEQWLAYWILYSFLTLSELILQSLLEWIPIWYTAKLVFVAW 61
Query: 353 LVLPWFNGAAYVYDHFVRPMF 415
LVLP F GAA++Y+ VR F
Sbjct: 62 LVLPQFRGAAFIYNKVVREQF 82
>sptr|Q9SW30|Q9SW30 Abscisic acid-induced-like protein.
Length = 116
Score = 104 bits (259), Expect = 2e-21
Identities = 45/81 (55%), Positives = 57/81 (70%)
Frame = +2
Query: 173 LAYPLYASVRAIETKNPVDDQQWLTYWVLYSFITLFELTFAPIIEWLPFWSYAKLFFNCW 352
L YPLYASV A+E+ VDD+QWL YW++YSF++L EL +IEW+P W KL F W
Sbjct: 2 LLYPLYASVIAMESTTKVDDEQWLAYWIIYSFLSLTELILQSLIEWIPIWYTVKLVFVAW 61
Query: 353 LVLPWFNGAAYVYDHFVRPMF 415
LVLP F GAA++Y+ VR F
Sbjct: 62 LVLPQFQGAAFIYNRVVREQF 82
>sptr|Q9UU91|Q9UU91 Putative transport protein.
Length = 182
Score = 79.0 bits (193), Expect = 1e-13
Identities = 36/92 (39%), Positives = 55/92 (59%)
Frame = +2
Query: 164 LISLAYPLYASVRAIETKNPVDDQQWLTYWVLYSFITLFELTFAPIIEWLPFWSYAKLFF 343
L++ A P + S+ AIET N DD QWLTY+++ SF+ + E I+ ++P + K F
Sbjct: 62 LLAFAMPAFFSINAIETTNKADDTQWLTYYLVTSFLNVIEYWSQLILYYVPVYWLLKAIF 121
Query: 344 NCWLVLPWFNGAAYVYDHFVRPMFVNRQIVNI 439
WL LP FNGA +Y H +RP ++ ++ I
Sbjct: 122 LIWLALPKFNGATIIYRHLIRP-YITPHVIRI 152
>sw|Q60870|DP1_MOUSE Polyposis locus protein 1 homolog (TB2 protein
homolog) (GP106).
Length = 185
Score = 76.6 bits (187), Expect = 5e-13
Identities = 36/89 (40%), Positives = 50/89 (56%), Gaps = 1/89 (1%)
Frame = +2
Query: 164 LISLAYPLYASVRAIETKNPVDDQQWLTYWVLYSFITLFELTFAPIIEWLPFWSYAKLFF 343
LI YP Y S++AIE+ N DD QWLTYWV+Y ++ E + WLPF+ K F
Sbjct: 57 LIGFGYPAYISMKAIESPNKDDDTQWLTYWVVYGVFSIAEFFSDLFLSWLPFYYMLKCGF 116
Query: 344 NCWLVLPW-FNGAAYVYDHFVRPMFVNRQ 427
W + P NGA +Y +RP+F+ +
Sbjct: 117 LLWCMAPSPANGAEMLYRRIIRPIFLRHE 145
>sw|Q00765|DP1_HUMAN Polyposis locus protein 1 (TB2 protein).
Length = 185
Score = 76.3 bits (186), Expect = 7e-13
Identities = 35/89 (39%), Positives = 48/89 (53%), Gaps = 1/89 (1%)
Frame = +2
Query: 164 LISLAYPLYASVRAIETKNPVDDQQWLTYWVLYSFITLFELTFAPIIEWLPFWSYAKLFF 343
LI YP Y S++AIE+ N DD QWLTYWV+Y ++ E + W PF+ K F
Sbjct: 57 LIGFGYPAYISIKAIESPNKEDDTQWLTYWVVYGVFSIAEFFSDIFLSWFPFYYMLKCGF 116
Query: 344 NCWLVLPW-FNGAAYVYDHFVRPMFVNRQ 427
W + P NGA +Y +RP F+ +
Sbjct: 117 LLWCMAPSPSNGAELLYKRIIRPFFLKHE 145
>sptr|Q9CQG4|Q9CQG4 Deleted in polyposis 1.
Length = 185
Score = 75.1 bits (183), Expect = 2e-12
Identities = 35/89 (39%), Positives = 49/89 (55%), Gaps = 1/89 (1%)
Frame = +2
Query: 164 LISLAYPLYASVRAIETKNPVDDQQWLTYWVLYSFITLFELTFAPIIEWLPFWSYAKLFF 343
LI YP Y S++AIE+ N DD QWLTYWV+Y ++ E + W PF+ K F
Sbjct: 57 LIGFGYPAYISMKAIESPNKDDDTQWLTYWVVYGVFSIAEFFSDLFLSWFPFYYMLKCGF 116
Query: 344 NCWLVLPW-FNGAAYVYDHFVRPMFVNRQ 427
W + P NGA +Y +RP+F+ +
Sbjct: 117 LLWCMAPSPANGAEMLYRRIIRPIFLKHE 145
>sptr|Q96LM0|Q96LM0 Hypothetical protein FLJ25383 (TB2 protein-like
1).
Length = 184
Score = 74.3 bits (181), Expect = 3e-12
Identities = 37/87 (42%), Positives = 49/87 (56%), Gaps = 2/87 (2%)
Frame = +2
Query: 164 LISLAYPLYASVRAIETKNPVDDQQWLTYWVLYSFITLFELTFAPIIEWLPFWSYAKLFF 343
LI YP YAS++AIE+ + DD WLTYWV+Y+ L E ++ W PF+ K F
Sbjct: 60 LIGFVYPAYASIKAIESPSKDDDTVWLTYWVVYALFGLAEFFSDLLLSWFPFYYVGKCAF 119
Query: 344 --NCWLVLPWFNGAAYVYDHFVRPMFV 418
C PW NGA +Y VRP+F+
Sbjct: 120 LLFCMAPRPW-NGALMLYQRVVRPLFL 145
>sptr|Q96HR9|Q96HR9 Hypothetical protein.
Length = 184
Score = 74.3 bits (181), Expect = 3e-12
Identities = 37/87 (42%), Positives = 49/87 (56%), Gaps = 2/87 (2%)
Frame = +2
Query: 164 LISLAYPLYASVRAIETKNPVDDQQWLTYWVLYSFITLFELTFAPIIEWLPFWSYAKLFF 343
LI YP YAS++AIE+ + DD WLTYWV+Y+ L E ++ W PF+ K F
Sbjct: 60 LIGFVYPAYASIKAIESPSKDDDTVWLTYWVVYALFGLAEFFSDLLLSWFPFYYVGKCAF 119
Query: 344 --NCWLVLPWFNGAAYVYDHFVRPMFV 418
C PW NGA +Y VRP+F+
Sbjct: 120 LLFCMAPRPW-NGALMLYQRVVRPLFL 145
>sptr|Q921E4|Q921E4 Deleted in polyposis 1.
Length = 185
Score = 73.6 bits (179), Expect = 4e-12
Identities = 35/89 (39%), Positives = 48/89 (53%), Gaps = 1/89 (1%)
Frame = +2
Query: 164 LISLAYPLYASVRAIETKNPVDDQQWLTYWVLYSFITLFELTFAPIIEWLPFWSYAKLFF 343
LI YP Y S++AIE+ N DD QWLTYWV+Y ++ E + W PF+ K F
Sbjct: 57 LIGFGYPAYISMKAIESPNKDDDTQWLTYWVVYGVFSIAEFFSDLFLSWFPFYYMLKCGF 116
Query: 344 NCWLVLPW-FNGAAYVYDHFVRPMFVNRQ 427
W + P NGA Y +RP+F+ +
Sbjct: 117 LLWCMAPSPANGAEMRYRRIIRPIFLKHE 145
>sptr|Q9N4G8|Q9N4G8 Hypothetical 20.6 kDa protein.
Length = 183
Score = 73.2 bits (178), Expect = 6e-12
Identities = 33/82 (40%), Positives = 49/82 (59%)
Frame = +2
Query: 167 ISLAYPLYASVRAIETKNPVDDQQWLTYWVLYSFITLFELTFAPIIEWLPFWSYAKLFFN 346
+ YP Y S++AIE+ N DD QWLTYWV+++ +++ E I+ P + K F
Sbjct: 67 MGFVYPAYMSIKAIESSNKEDDTQWLTYWVIFAILSVVEFFSVQIVAVFPVYWLFKSIFL 126
Query: 347 CWLVLPWFNGAAYVYDHFVRPM 412
+L LP F GAA +Y FV+P+
Sbjct: 127 LYLYLPSFLGAAKLYHRFVKPV 148
>sptr|O77388|O77388 Hypothetical 26.0 kDa protein.
Length = 221
Score = 73.2 bits (178), Expect = 6e-12
Identities = 30/88 (34%), Positives = 52/88 (59%)
Frame = +2
Query: 164 LISLAYPLYASVRAIETKNPVDDQQWLTYWVLYSFITLFELTFAPIIEWLPFWSYAKLFF 343
++ AYP Y S +A+E+++ + + WLTYWV++S E I+ W+PF+ KL F
Sbjct: 95 VVGFAYPAYQSFKAVESQSRDETKLWLTYWVVFSLFFFIEYLIDIILFWVPFYYLIKLLF 154
Query: 344 NCWLVLPWFNGAAYVYDHFVRPMFVNRQ 427
+L +P GA VY++ +RP+ + +
Sbjct: 155 LLYLYMPQVRGAVMVYNYIIRPILLKHE 182
>sptrnew|EAA05239|EAA05239 ENSANGP00000018512 (Fragment).
Length = 321
Score = 72.8 bits (177), Expect = 8e-12
Identities = 34/100 (34%), Positives = 54/100 (54%)
Frame = +2
Query: 128 VLAKNFDVLAGPLISLAYPLYASVRAIETKNPVDDQQWLTYWVLYSFITLFELTFAPIIE 307
+L F +L G L YP YAS +A+ TKN + +W+ YW++++F T E ++
Sbjct: 69 ILFPGFRLLFGTL----YPAYASYKAVRTKNVKEYVKWMMYWIVFAFFTFIETFTDILLS 124
Query: 308 WLPFWSYAKLFFNCWLVLPWFNGAAYVYDHFVRPMFVNRQ 427
W PF+ K+ WL+ P G++ +Y FV PM R+
Sbjct: 125 WFPFYYEIKVIVVLWLLSPATRGSSTLYRKFVHPMLTRRE 164
>sptrnew|AAH06218|AAH06218 Hypothetical protein.
Length = 252
Score = 72.4 bits (176), Expect = 1e-11
Identities = 28/83 (33%), Positives = 50/83 (60%)
Frame = +2
Query: 179 YPLYASVRAIETKNPVDDQQWLTYWVLYSFITLFELTFAPIIEWLPFWSYAKLFFNCWLV 358
YP Y+S +A++TKN + +W+ YW++++F T E ++ W PF+ K+ F WL+
Sbjct: 18 YPAYSSYKAVKTKNVKEYVKWMMYWIVFAFFTTAETLTDIVLSWFPFYFELKIAFVIWLL 77
Query: 359 LPWFNGAAYVYDHFVRPMFVNRQ 427
P+ G++ +Y FV P N++
Sbjct: 78 SPYTKGSSVLYRKFVHPTLSNKE 100
>sptr|Q9JM62|Q9JM62 Polyposis locus protein 1-like 1 (TB2
protein-like 1).
Length = 201
Score = 71.6 bits (174), Expect = 2e-11
Identities = 36/91 (39%), Positives = 50/91 (54%), Gaps = 2/91 (2%)
Frame = +2
Query: 164 LISLAYPLYASVRAIETKNPVDDQQWLTYWVLYSFITLFELTFAPIIEWLPFWSYAKLFF 343
+I YP YASV+AIE+ + DD WLTYWV+Y+ L E ++ W PF+ K F
Sbjct: 60 VIGFVYPAYASVKAIESPSKEDDTVWLTYWVVYALFGLVEFFSDLLLFWFPFYYAGKCAF 119
Query: 344 --NCWLVLPWFNGAAYVYDHFVRPMFVNRQI 430
C PW NGA +Y +RP+F+ +
Sbjct: 120 LLFCMTPGPW-NGALLLYHRVIRPLFLKHHM 149
>sptr|Q8K0X2|Q8K0X2 Hypothetical protein.
Length = 174
Score = 71.6 bits (174), Expect = 2e-11
Identities = 36/91 (39%), Positives = 50/91 (54%), Gaps = 2/91 (2%)
Frame = +2
Query: 164 LISLAYPLYASVRAIETKNPVDDQQWLTYWVLYSFITLFELTFAPIIEWLPFWSYAKLFF 343
+I YP YASV+AIE+ + DD WLTYWV+Y+ L E ++ W PF+ K F
Sbjct: 60 VIGFVYPAYASVKAIESPSKEDDTVWLTYWVVYALFGLVEFFSDLLLFWFPFYYAGKCAF 119
Query: 344 --NCWLVLPWFNGAAYVYDHFVRPMFVNRQI 430
C PW NGA +Y +RP+F+ +
Sbjct: 120 LLFCMTPGPW-NGALLLYHRVIRPLFLKHHM 149
>sptrnew|EAA31247|EAA31247 Hypothetical protein.
Length = 385
Score = 71.2 bits (173), Expect = 2e-11
Identities = 39/129 (30%), Positives = 67/129 (51%), Gaps = 3/129 (2%)
Frame = +2
Query: 143 FDVLAGPLISLA---YPLYASVRAIETKNPVDDQQWLTYWVLYSFITLFELTFAPIIEWL 313
FD+L L S+A +P+YAS +A+ + +P WL YWV+ SF+ L E + W+
Sbjct: 2 FDILPNLLSSVASFLFPVYASYKALRSSDPAQLTPWLMYWVVISFVVLVESWVGWFLVWV 61
Query: 314 PFWSYAKLFFNCWLVLPWFNGAAYVYDHFVRPMFVNRQIVNIWYIPRNEKLGKSDDVLSA 493
PF+++ + F +LVLP GA +Y + P ++ NE G+ +D +++
Sbjct: 62 PFYAFMRFVFLLYLVLPQTQGARVIYQTHIEP-----------WLEANE--GQIEDFIAS 108
Query: 494 AERYIEQNG 520
A ++ G
Sbjct: 109 AHERLKAAG 117
>sptrnew|EAA13329|EAA13329 ENSANGP00000003353 (Fragment).
Length = 143
Score = 70.9 bits (172), Expect = 3e-11
Identities = 31/85 (36%), Positives = 53/85 (62%), Gaps = 1/85 (1%)
Frame = +2
Query: 164 LISLAYPLYASVRAIETKNPVDDQQWLTYWVLYSFITLFELTFAPIIEWLPFWSYAKLFF 343
+I +AYP Y S++AIET+ DD +WLTYWV + +++FE +++ +PF+ K F
Sbjct: 54 VIGVAYPAYISMKAIETRTKEDDTKWLTYWVTFGVLSVFEHFSFFLVQIIPFYWLLKCLF 113
Query: 344 NCWLVLPW-FNGAAYVYDHFVRPMF 415
+ W ++P NG+ +Y ++P F
Sbjct: 114 HIWCMVPMENNGSTIMYHKVIQPYF 138
>sptrnew|EAA13321|EAA13321 AgCP2771 (Fragment).
Length = 164
Score = 70.9 bits (172), Expect = 3e-11
Identities = 31/85 (36%), Positives = 53/85 (62%), Gaps = 1/85 (1%)
Frame = +2
Query: 164 LISLAYPLYASVRAIETKNPVDDQQWLTYWVLYSFITLFELTFAPIIEWLPFWSYAKLFF 343
+I +AYP Y S++AIET+ DD +WLTYWV + +++FE +++ +PF+ K F
Sbjct: 48 VIGVAYPAYISMKAIETRTKEDDTKWLTYWVTFGVLSVFEHFSFFLVQIIPFYWLLKCLF 107
Query: 344 NCWLVLPW-FNGAAYVYDHFVRPMF 415
+ W ++P NG+ +Y ++P F
Sbjct: 108 HIWCMVPMENNGSTIMYHKVIQPYF 132
>sptr|O45108|O45108 Hypothetical 22.8 kDa protein.
Length = 205
Score = 68.9 bits (167), Expect = 1e-10
Identities = 34/96 (35%), Positives = 50/96 (52%), Gaps = 2/96 (2%)
Frame = +2
Query: 164 LISLAYPLYASVRAIETKNPVDDQQWLTYWVLYSFITLFELTFAPIIEWLPFWSYAKLFF 343
LI YP YASV+AI + DD WL YW ++ + L + I+ W PF+ AK F
Sbjct: 106 LIGFGYPTYASVKAIRSPGGDDDTVWLIYWTCFAVLYLVDFFSEAILSWFPFYYIAKACF 165
Query: 344 NCWLVLPWFNGAAYVYDHFVRPM--FVNRQIVNIWY 445
+L LP G+ Y+ V P+ FV++ + +Y
Sbjct: 166 LVYLYLPQTQGSVMFYETIVDPLVIFVDKNLDKFYY 201
>sptr|O93846|O93846 Pathogenicity protein.
Length = 369
Score = 68.6 bits (166), Expect = 1e-10
Identities = 33/92 (35%), Positives = 53/92 (57%), Gaps = 3/92 (3%)
Frame = +2
Query: 143 FDVLAGPLISLA---YPLYASVRAIETKNPVDDQQWLTYWVLYSFITLFELTFAPIIEWL 313
FD+ A L S+A +PL+AS +A++T +P WL YWV+ + L E + W+
Sbjct: 2 FDIFAKLLSSIASFLFPLFASYKALKTTDPAQLTPWLMYWVVLACALLVESWTEWFLCWI 61
Query: 314 PFWSYAKLFFNCWLVLPWFNGAAYVYDHFVRP 409
PF++Y +L F +L+LP GA +Y ++ P
Sbjct: 62 PFYAYIRLLFLLYLILPQTQGARVLYQDYIHP 93
>sptr|Q8MLS9|Q8MLS9 CG30193-PA (RE29641p).
Length = 288
Score = 68.2 bits (165), Expect = 2e-10
Identities = 32/99 (32%), Positives = 53/99 (53%)
Frame = +2
Query: 179 YPLYASVRAIETKNPVDDQQWLTYWVLYSFITLFELTFAPIIEWLPFWSYAKLFFNCWLV 358
YP YAS +A+ TK+ + +W+ YW++++F T E I WLPF+ K+ WL+
Sbjct: 18 YPAYASYKAVRTKDVKEYVKWMMYWIVFAFFTCIETFTDIFISWLPFYYEVKVALVFWLL 77
Query: 359 LPWFNGAAYVYDHFVRPMFVNRQIVNIWYIPRNEKLGKS 475
P G++ +Y FV PM + Y+ + ++ G S
Sbjct: 78 SPATKGSSTLYRKFVHPMLTRHEQEIDEYVNQAKERGYS 116
Database: /db/trembl-ebi/tmp/swall
Posted date: Sep 26, 2003 8:29 PM
Number of letters in database: 406,886,216
Number of sequences in database: 1,272,877
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 717,167,226
Number of Sequences: 1272877
Number of extensions: 14539971
Number of successful extensions: 42063
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 39407
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 41970
length of database: 406,886,216
effective HSP length: 123
effective length of database: 250,322,345
effective search space used: 56072205280
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

