BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= 2750560.2.1
(660 letters)
Database: /db/trembl-ebi/tmp/swall
1,302,759 sequences; 415,658,970 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
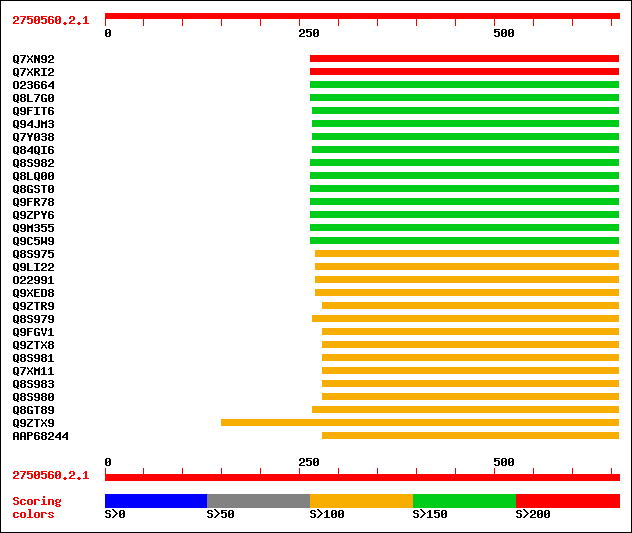
sptr|Q7XN92|Q7XN92 OSJNBa0064D20.11 protein. 216 2e-55
sptr|Q7XRI2|Q7XRI2 P0076O17.10 protein. 216 2e-55
sptr|O23664|O23664 Auxin response factor 1. 188 6e-47
sptr|Q8L7G0|Q8L7G0 Auxin response factor 1. 185 5e-46
sptr|Q9FIT6|Q9FIT6 Auxin response factor-like protein. 182 4e-45
sptr|Q94JM3|Q94JM3 AT5g62000/mtg10_20. 182 4e-45
sptr|Q7Y038|Q7Y038 Auxin response factor-like protein. 179 3e-44
sptr|Q84QI6|Q84QI6 Auxin response factor-like protein. 179 3e-44
sptr|Q8S982|Q8S982 Auxin response factor 2 (Fragment). 176 3e-43
sptr|Q8LQ00|Q8LQ00 Auxin response factor 2. 176 3e-43
sptr|Q8GST0|Q8GST0 Auxin response factor 1. 171 1e-41
sptr|Q9FR78|Q9FR78 Putative auxin response factor 1. 166 2e-40
sptr|Q9ZPY6|Q9ZPY6 Putative ARF1 family auxin responsive transcr... 165 5e-40
sptr|Q9M355|Q9M355 Auxin response factor-like protein. 159 2e-38
sptr|Q9C5W9|Q9C5W9 ARF16 (Putative auxin response factor protein... 159 2e-38
sptr|Q8S975|Q8S975 Auxin response factor 16 (Fragment). 148 5e-35
sptr|Q9LI22|Q9LI22 EST C26826(C50159) corresponds to a region of... 148 5e-35
sptr|O22991|O22991 Auxin inducible protein isolog. 147 2e-34
sptr|Q9XED8|Q9XED8 Auxin response factor 9 (ARF9). 147 2e-34
sptr|Q9ZTR9|Q9ZTR9 Auxin response factor 8. 132 5e-30
sptr|Q8S979|Q8S979 Auxin response factor 7a (Fragment). 132 5e-30
sptr|Q9FGV1|Q9FGV1 Auxin responsive transcription factor. 132 5e-30
sptr|Q9ZTX8|Q9ZTX8 ARF6 (Auxin response factor 6). 131 9e-30
sptr|Q8S981|Q8S981 Auxin response factor 6a (Fragment). 130 1e-29
sptr|Q7XM11|Q7XM11 OSJNBa0084K01.20 protein. 128 6e-29
sptr|Q8S983|Q8S983 Arabidopsis Monopteros-like protein. 128 6e-29
sptr|Q8S980|Q8S980 Auxin response factor 6b (Fragment). 127 2e-28
sptr|Q8GT89|Q8GT89 Hypothetical transcription factor. 126 2e-28
sptr|Q9ZTX9|Q9ZTX9 Auxin response factor 4 (Auxin response facto... 126 2e-28
sptrnew|AAP68244|AAP68244 At1g19850. 126 3e-28
>sptr|Q7XN92|Q7XN92 OSJNBa0064D20.11 protein.
Length = 1134
Score = 216 bits (550), Expect = 2e-55
Identities = 104/132 (78%), Positives = 112/132 (84%)
Frame = +3
Query: 264 DALFRELWHACAGPLVTVPRQGELVYYFPQGHMEQLEASTDQQLDQHLPLFDLPPKILCK 443
DALFRELWHACAGPLVTVP++GE VYYFPQGHMEQLEAST+QQLDQ+LP+F+LP KILC
Sbjct: 15 DALFRELWHACAGPLVTVPKRGERVYYFPQGHMEQLEASTNQQLDQYLPMFNLPSKILCS 74
Query: 444 VVNVELRAETDSDEVYAQIMLQPEADQXXXXXXXXXXXXXXRCNVYSFCKTLTASDTSTH 623
VVNVELRAE DSDEVYAQIMLQPEADQ +C +SFCKTLTASDTSTH
Sbjct: 75 VVNVELRAEADSDEVYAQIMLQPEADQSELTSLDPELQDLEKCTAHSFCKTLTASDTSTH 134
Query: 624 GGFSVLRRHAEE 659
GGFSVLRRHAEE
Sbjct: 135 GGFSVLRRHAEE 146
>sptr|Q7XRI2|Q7XRI2 P0076O17.10 protein.
Length = 1673
Score = 216 bits (550), Expect = 2e-55
Identities = 104/132 (78%), Positives = 112/132 (84%)
Frame = +3
Query: 264 DALFRELWHACAGPLVTVPRQGELVYYFPQGHMEQLEASTDQQLDQHLPLFDLPPKILCK 443
DALFRELWHACAGPLVTVP++GE VYYFPQGHMEQLEAST+QQLDQ+LP+F+LP KILC
Sbjct: 15 DALFRELWHACAGPLVTVPKRGERVYYFPQGHMEQLEASTNQQLDQYLPMFNLPSKILCS 74
Query: 444 VVNVELRAETDSDEVYAQIMLQPEADQXXXXXXXXXXXXXXRCNVYSFCKTLTASDTSTH 623
VVNVELRAE DSDEVYAQIMLQPEADQ +C +SFCKTLTASDTSTH
Sbjct: 75 VVNVELRAEADSDEVYAQIMLQPEADQSELTSLDPELQDLEKCTAHSFCKTLTASDTSTH 134
Query: 624 GGFSVLRRHAEE 659
GGFSVLRRHAEE
Sbjct: 135 GGFSVLRRHAEE 146
>sptr|O23664|O23664 Auxin response factor 1.
Length = 665
Score = 188 bits (477), Expect = 6e-47
Identities = 89/132 (67%), Positives = 104/132 (78%)
Frame = +3
Query: 264 DALFRELWHACAGPLVTVPRQGELVYYFPQGHMEQLEASTDQQLDQHLPLFDLPPKILCK 443
DAL RELWHACAGPLVT+PR+GE VYYFP+GHMEQLEAS Q L+Q +P F+LP KILCK
Sbjct: 17 DALCRELWHACAGPLVTLPREGERVYYFPEGHMEQLEASMHQGLEQQMPSFNLPSKILCK 76
Query: 444 VVNVELRAETDSDEVYAQIMLQPEADQXXXXXXXXXXXXXXRCNVYSFCKTLTASDTSTH 623
V+N++ RAE ++DEVYAQI L PE DQ +C V+SFCKTLTASDTSTH
Sbjct: 77 VINIQRRAEPETDEVYAQITLLPELDQSEPTSPDAPVQEPEKCTVHSFCKTLTASDTSTH 136
Query: 624 GGFSVLRRHAEE 659
GGFSVLRRHA++
Sbjct: 137 GGFSVLRRHADD 148
>sptr|Q8L7G0|Q8L7G0 Auxin response factor 1.
Length = 662
Score = 185 bits (469), Expect = 5e-46
Identities = 88/132 (66%), Positives = 103/132 (78%)
Frame = +3
Query: 264 DALFRELWHACAGPLVTVPRQGELVYYFPQGHMEQLEASTDQQLDQHLPLFDLPPKILCK 443
DAL RELWHACAGPLVT+PR+GE VYYFP+GHMEQLEAS Q L+Q +P F+LP KILCK
Sbjct: 17 DALCRELWHACAGPLVTLPREGERVYYFPEGHMEQLEASMHQGLEQQMPSFNLPSKILCK 76
Query: 444 VVNVELRAETDSDEVYAQIMLQPEADQXXXXXXXXXXXXXXRCNVYSFCKTLTASDTSTH 623
V+N++ RAE ++DEVYAQI L PE DQ +C V+SFCKTLTASDTST
Sbjct: 77 VINIQRRAEPETDEVYAQITLLPELDQSEPTSPDAPVQEPEKCTVHSFCKTLTASDTSTQ 136
Query: 624 GGFSVLRRHAEE 659
GGFSVLRRHA++
Sbjct: 137 GGFSVLRRHADD 148
>sptr|Q9FIT6|Q9FIT6 Auxin response factor-like protein.
Length = 859
Score = 182 bits (461), Expect = 4e-45
Identities = 88/132 (66%), Positives = 106/132 (80%), Gaps = 1/132 (0%)
Frame = +3
Query: 267 ALFRELWHACAGPLVTVPRQGELVYYFPQGHMEQLEASTDQQLDQHLPLFDLPPKILCKV 446
AL+RELWHACAGPLVTVPRQ + V+YFPQGH+EQ+EAST+Q +Q +PL+DLP K+LC+V
Sbjct: 57 ALYRELWHACAGPLVTVPRQDDRVFYFPQGHIEQVEASTNQAAEQQMPLYDLPSKLLCRV 116
Query: 447 VNVELRAETDSDEVYAQIMLQPEADQXXXXXXXXX-XXXXXRCNVYSFCKTLTASDTSTH 623
+NV+L+AE D+DEVYAQI L PEA+Q R V+SFCKTLTASDTSTH
Sbjct: 117 INVDLKAEADTDEVYAQITLLPEANQDENAIEKEAPLPPPPRFQVHSFCKTLTASDTSTH 176
Query: 624 GGFSVLRRHAEE 659
GGFSVLRRHA+E
Sbjct: 177 GGFSVLRRHADE 188
>sptr|Q94JM3|Q94JM3 AT5g62000/mtg10_20.
Length = 678
Score = 182 bits (461), Expect = 4e-45
Identities = 88/132 (66%), Positives = 106/132 (80%), Gaps = 1/132 (0%)
Frame = +3
Query: 267 ALFRELWHACAGPLVTVPRQGELVYYFPQGHMEQLEASTDQQLDQHLPLFDLPPKILCKV 446
AL+RELWHACAGPLVTVPRQ + V+YFPQGH+EQ+EAST+Q +Q +PL+DLP K+LC+V
Sbjct: 57 ALYRELWHACAGPLVTVPRQDDRVFYFPQGHIEQVEASTNQAAEQQMPLYDLPSKLLCRV 116
Query: 447 VNVELRAETDSDEVYAQIMLQPEADQXXXXXXXXX-XXXXXRCNVYSFCKTLTASDTSTH 623
+NV+L+AE D+DEVYAQI L PEA+Q R V+SFCKTLTASDTSTH
Sbjct: 117 INVDLKAEADTDEVYAQITLLPEANQDENAIEKEAPLPPPPRFQVHSFCKTLTASDTSTH 176
Query: 624 GGFSVLRRHAEE 659
GGFSVLRRHA+E
Sbjct: 177 GGFSVLRRHADE 188
>sptr|Q7Y038|Q7Y038 Auxin response factor-like protein.
Length = 326
Score = 179 bits (454), Expect = 3e-44
Identities = 88/132 (66%), Positives = 106/132 (80%), Gaps = 1/132 (0%)
Frame = +3
Query: 267 ALFRELWHACAGPLVTVPRQGELVYYFPQGHMEQLEASTDQQLDQHLPLFDLPPKILCKV 446
AL++ELWHACAGPLVTVPRQGE VYYFPQGH+EQ+EAST+Q DQ +P++DL KILC+V
Sbjct: 33 ALYKELWHACAGPLVTVPRQGERVYYFPQGHIEQVEASTNQFADQQMPIYDLRSKILCRV 92
Query: 447 VNVELRAETDSDEVYAQIMLQPEADQXXXXXXXX-XXXXXXRCNVYSFCKTLTASDTSTH 623
+NV+L+A+ D+DEV+AQI L PE +Q R +V+SFCKTLTASDTSTH
Sbjct: 93 INVQLKAKPDTDEVFAQITLLPEPNQDENAVEKEPPPPLLPRFHVHSFCKTLTASDTSTH 152
Query: 624 GGFSVLRRHAEE 659
GGFSVLRRHAEE
Sbjct: 153 GGFSVLRRHAEE 164
>sptr|Q84QI6|Q84QI6 Auxin response factor-like protein.
Length = 840
Score = 179 bits (454), Expect = 3e-44
Identities = 88/132 (66%), Positives = 106/132 (80%), Gaps = 1/132 (0%)
Frame = +3
Query: 267 ALFRELWHACAGPLVTVPRQGELVYYFPQGHMEQLEASTDQQLDQHLPLFDLPPKILCKV 446
AL++ELWHACAGPLVTVPRQGE VYYFPQGH+EQ+EAST+Q DQ +P++DL KILC+V
Sbjct: 33 ALYKELWHACAGPLVTVPRQGERVYYFPQGHIEQVEASTNQFADQQMPIYDLRSKILCRV 92
Query: 447 VNVELRAETDSDEVYAQIMLQPEADQXXXXXXXX-XXXXXXRCNVYSFCKTLTASDTSTH 623
+NV+L+A+ D+DEV+AQI L PE +Q R +V+SFCKTLTASDTSTH
Sbjct: 93 INVQLKAKPDTDEVFAQITLLPEPNQDENAVEKEPPPPLLPRFHVHSFCKTLTASDTSTH 152
Query: 624 GGFSVLRRHAEE 659
GGFSVLRRHAEE
Sbjct: 153 GGFSVLRRHAEE 164
>sptr|Q8S982|Q8S982 Auxin response factor 2 (Fragment).
Length = 791
Score = 176 bits (445), Expect = 3e-43
Identities = 86/135 (63%), Positives = 102/135 (75%), Gaps = 3/135 (2%)
Frame = +3
Query: 264 DALFRELWHACAGPLVTVPRQGELVYYFPQGHMEQLEASTDQQLDQHLPLFDLPPKILCK 443
D L+ ELWHACAGPLVTVPR G+LV+YFPQGH+EQ+EAS +Q D + L+DLP K+LC+
Sbjct: 2 DPLYDELWHACAGPLVTVPRVGDLVFYFPQGHIEQVEASMNQVADSQMRLYDLPSKLLCR 61
Query: 444 VVNVELRAETDSDEVYAQIMLQPEADQXXXXXXXXXXXX---XXRCNVYSFCKTLTASDT 614
V+NVEL+AE D+DEVYAQ+ML PE +Q R V SFCKTLTASDT
Sbjct: 62 VLNVELKAEQDTDEVYAQVMLMPEPEQNEMAVEKTTPTSGPVQARPPVRSFCKTLTASDT 121
Query: 615 STHGGFSVLRRHAEE 659
STHGGFSVLRRHA+E
Sbjct: 122 STHGGFSVLRRHADE 136
>sptr|Q8LQ00|Q8LQ00 Auxin response factor 2.
Length = 826
Score = 176 bits (445), Expect = 3e-43
Identities = 86/135 (63%), Positives = 102/135 (75%), Gaps = 3/135 (2%)
Frame = +3
Query: 264 DALFRELWHACAGPLVTVPRQGELVYYFPQGHMEQLEASTDQQLDQHLPLFDLPPKILCK 443
D L+ ELWHACAGPLVTVPR G+LV+YFPQGH+EQ+EAS +Q D + L+DLP K+LC+
Sbjct: 14 DPLYDELWHACAGPLVTVPRVGDLVFYFPQGHIEQVEASMNQVADSQMRLYDLPSKLLCR 73
Query: 444 VVNVELRAETDSDEVYAQIMLQPEADQXXXXXXXXXXXX---XXRCNVYSFCKTLTASDT 614
V+NVEL+AE D+DEVYAQ+ML PE +Q R V SFCKTLTASDT
Sbjct: 74 VLNVELKAEQDTDEVYAQVMLMPEPEQNEMAVEKTTPTSGPVQARPPVRSFCKTLTASDT 133
Query: 615 STHGGFSVLRRHAEE 659
STHGGFSVLRRHA+E
Sbjct: 134 STHGGFSVLRRHADE 148
>sptr|Q8GST0|Q8GST0 Auxin response factor 1.
Length = 836
Score = 171 bits (432), Expect = 1e-41
Identities = 87/138 (63%), Positives = 103/138 (74%), Gaps = 6/138 (4%)
Frame = +3
Query: 264 DALFRELWHACAGPLVTVPRQGELVYYFPQGHMEQLEASTDQQLDQHLPLFDLPPKILCK 443
DALF ELW ACAGPLVTVPR GE V+YFPQGH+EQ+EAST+Q +Q + L++LP KILC+
Sbjct: 19 DALFTELWSACAGPLVTVPRVGEKVFYFPQGHIEQVEASTNQVGEQRMQLYNLPWKILCE 78
Query: 444 VVNVELRAETDSDEVYAQIMLQPEADQXXXXXX------XXXXXXXXRCNVYSFCKTLTA 605
V+NVEL+AE D+DEVYAQ+ L PE+ Q R V+SFCKTLTA
Sbjct: 79 VMNVELKAEPDTDEVYAQLTLLPESKQQEDNGSTEEEVPSAPAAGHVRPRVHSFCKTLTA 138
Query: 606 SDTSTHGGFSVLRRHAEE 659
SDTSTHGGFSVLRRHA+E
Sbjct: 139 SDTSTHGGFSVLRRHADE 156
>sptr|Q9FR78|Q9FR78 Putative auxin response factor 1.
Length = 857
Score = 166 bits (420), Expect = 2e-40
Identities = 85/138 (61%), Positives = 101/138 (73%), Gaps = 6/138 (4%)
Frame = +3
Query: 264 DALFRELWHACAGPLVTVPRQGELVYYFPQGHMEQLEASTDQQLDQHLPLFDLPPKILCK 443
DALF ELW ACAGPLVTVPR GE +YFPQGH+EQ+EAST+Q +Q + L++LP KILC+
Sbjct: 35 DALFTELWSACAGPLVTVPRVGEKEFYFPQGHIEQVEASTNQVGEQRMQLYNLPWKILCE 94
Query: 444 VVNVELRAETDSDEVYAQIMLQPEADQXXXXXX------XXXXXXXXRCNVYSFCKTLTA 605
V+NVEL+AE D+DEVYAQ+ L PE + R V+SFCKTLTA
Sbjct: 95 VMNVELKAEPDTDEVYAQLTLLPELKRQEDNGSTEEEVPSAPAAGHVRPRVHSFCKTLTA 154
Query: 606 SDTSTHGGFSVLRRHAEE 659
SDTSTHGGFSVLRRHA+E
Sbjct: 155 SDTSTHGGFSVLRRHADE 172
>sptr|Q9ZPY6|Q9ZPY6 Putative ARF1 family auxin responsive
transcription factor.
Length = 622
Score = 165 bits (417), Expect = 5e-40
Identities = 84/133 (63%), Positives = 99/133 (74%), Gaps = 1/133 (0%)
Frame = +3
Query: 264 DALFRELWHACAGPLVTVPRQGELVYYFPQGHMEQLEASTDQQL-DQHLPLFDLPPKILC 440
D L+ ELW ACAGPLV VPR GE V+YFPQGHMEQL AST+Q + DQ +P+F+LPPKILC
Sbjct: 37 DELYTELWKACAGPLVEVPRYGERVFYFPQGHMEQLVASTNQGVVDQEIPVFNLPPKILC 96
Query: 441 KVVNVELRAETDSDEVYAQIMLQPEADQXXXXXXXXXXXXXXRCNVYSFCKTLTASDTST 620
+V++V L+AE ++DEVYAQI LQPE DQ + V SF K LTASDTST
Sbjct: 97 RVLSVTLKAEHETDEVYAQITLQPEEDQSEPTSLDPPLVEPAKPTVDSFVKILTASDTST 156
Query: 621 HGGFSVLRRHAEE 659
HGGFSVLR+HA E
Sbjct: 157 HGGFSVLRKHATE 169
>sptr|Q9M355|Q9M355 Auxin response factor-like protein.
Length = 613
Score = 159 bits (403), Expect = 2e-38
Identities = 80/133 (60%), Positives = 97/133 (72%), Gaps = 1/133 (0%)
Frame = +3
Query: 264 DALFRELWHACAGPLVTVPRQGELVYYFPQGHMEQLEASTDQQLD-QHLPLFDLPPKILC 440
D L+ ELW CAGPLV VPR E V+YFPQGHMEQL AST+Q ++ + +P+FDLPPKILC
Sbjct: 20 DQLYTELWKVCAGPLVEVPRAQERVFYFPQGHMEQLVASTNQGINSEEIPVFDLPPKILC 79
Query: 441 KVVNVELRAETDSDEVYAQIMLQPEADQXXXXXXXXXXXXXXRCNVYSFCKTLTASDTST 620
+V++V L+AE ++DEVYAQI LQPE DQ + +SF K LTASDTST
Sbjct: 80 RVLDVTLKAEHETDEVYAQITLQPEEDQSEPTSLDPPIVGPTKQEFHSFVKILTASDTST 139
Query: 621 HGGFSVLRRHAEE 659
HGGFSVLR+HA E
Sbjct: 140 HGGFSVLRKHATE 152
>sptr|Q9C5W9|Q9C5W9 ARF16 (Putative auxin response factor protein)
(AT3g61830/F15G16_220).
Length = 602
Score = 159 bits (403), Expect = 2e-38
Identities = 80/133 (60%), Positives = 97/133 (72%), Gaps = 1/133 (0%)
Frame = +3
Query: 264 DALFRELWHACAGPLVTVPRQGELVYYFPQGHMEQLEASTDQQLD-QHLPLFDLPPKILC 440
D L+ ELW CAGPLV VPR E V+YFPQGHMEQL AST+Q ++ + +P+FDLPPKILC
Sbjct: 20 DQLYTELWKVCAGPLVEVPRAQERVFYFPQGHMEQLVASTNQGINSEEIPVFDLPPKILC 79
Query: 441 KVVNVELRAETDSDEVYAQIMLQPEADQXXXXXXXXXXXXXXRCNVYSFCKTLTASDTST 620
+V++V L+AE ++DEVYAQI LQPE DQ + +SF K LTASDTST
Sbjct: 80 RVLDVTLKAEHETDEVYAQITLQPEEDQSEPTSLDPPIVGPTKQEFHSFVKILTASDTST 139
Query: 621 HGGFSVLRRHAEE 659
HGGFSVLR+HA E
Sbjct: 140 HGGFSVLRKHATE 152
>sptr|Q8S975|Q8S975 Auxin response factor 16 (Fragment).
Length = 695
Score = 148 bits (374), Expect = 5e-35
Identities = 79/132 (59%), Positives = 94/132 (71%), Gaps = 2/132 (1%)
Frame = +3
Query: 270 LFRELWHACAGPLVTVPRQGELVYYFPQGHMEQLEASTDQQL-DQHLPLFDLPPKILCKV 446
LF ELW ACAGPLV VP++ E V+YF QGH+EQL+ TD L + + +F +P KILCKV
Sbjct: 11 LFAELWRACAGPLVEVPQRDERVFYFLQGHLEQLQEPTDPALLAEQIKMFQVPYKILCKV 70
Query: 447 VNVELRAETDSDEVYAQIMLQPEADQXXX-XXXXXXXXXXXRCNVYSFCKTLTASDTSTH 623
VNVEL+AET++DEV+AQI LQP+ DQ R V+SFCK LT SDTSTH
Sbjct: 71 VNVELKAETETDEVFAQITLQPDPDQENLPTLPDPPLPEQPRPVVHSFCKILTPSDTSTH 130
Query: 624 GGFSVLRRHAEE 659
GGFSVLRRHA E
Sbjct: 131 GGFSVLRRHANE 142
>sptr|Q9LI22|Q9LI22 EST C26826(C50159) corresponds to a region of
the predicted gene.
Length = 613
Score = 148 bits (374), Expect = 5e-35
Identities = 79/132 (59%), Positives = 94/132 (71%), Gaps = 2/132 (1%)
Frame = +3
Query: 270 LFRELWHACAGPLVTVPRQGELVYYFPQGHMEQLEASTDQQL-DQHLPLFDLPPKILCKV 446
LF ELW ACAGPLV VP++ E V+YF QGH+EQL+ TD L + + +F +P KILCKV
Sbjct: 15 LFAELWRACAGPLVEVPQRDERVFYFLQGHLEQLQEPTDPALLAEQIKMFQVPYKILCKV 74
Query: 447 VNVELRAETDSDEVYAQIMLQPEADQXXX-XXXXXXXXXXXRCNVYSFCKTLTASDTSTH 623
VNVEL+AET++DEV+AQI LQP+ DQ R V+SFCK LT SDTSTH
Sbjct: 75 VNVELKAETETDEVFAQITLQPDPDQENLPTLPDPPLPEQPRPVVHSFCKILTPSDTSTH 134
Query: 624 GGFSVLRRHAEE 659
GGFSVLRRHA E
Sbjct: 135 GGFSVLRRHANE 146
>sptr|O22991|O22991 Auxin inducible protein isolog.
Length = 497
Score = 147 bits (370), Expect = 2e-34
Identities = 83/134 (61%), Positives = 92/134 (68%), Gaps = 4/134 (2%)
Frame = +3
Query: 270 LFRELWHACAGPLVTVPRQGELVYYFPQGHMEQLEASTDQ-QLDQHLPLFDLPPKILCKV 446
L+ ELW CAGPLV VP+ E VYYFPQGHMEQLEAST Q L+ PLF LPPKILC V
Sbjct: 44 LYDELWKLCAGPLVDVPQAQERVYYFPQGHMEQLEASTQQVDLNTMKPLFVLPPKILCNV 103
Query: 447 VNVELRAETDSDEVYAQIMLQP---EADQXXXXXXXXXXXXXXRCNVYSFCKTLTASDTS 617
+NV L+AE D+DEVYAQI L P E D+ R V+SF K LTASDTS
Sbjct: 104 MNVSLQAEKDTDEVYAQITLIPVGTEVDE--PMSPDPSPPELQRPKVHSFSKVLTASDTS 161
Query: 618 THGGFSVLRRHAEE 659
THGGFSVLR+HA E
Sbjct: 162 THGGFSVLRKHATE 175
>sptr|Q9XED8|Q9XED8 Auxin response factor 9 (ARF9).
Length = 638
Score = 147 bits (370), Expect = 2e-34
Identities = 83/134 (61%), Positives = 92/134 (68%), Gaps = 4/134 (2%)
Frame = +3
Query: 270 LFRELWHACAGPLVTVPRQGELVYYFPQGHMEQLEASTDQ-QLDQHLPLFDLPPKILCKV 446
L+ ELW CAGPLV VP+ E VYYFPQGHMEQLEAST Q L+ PLF LPPKILC V
Sbjct: 9 LYDELWKLCAGPLVDVPQAQERVYYFPQGHMEQLEASTQQVDLNTMKPLFVLPPKILCNV 68
Query: 447 VNVELRAETDSDEVYAQIMLQP---EADQXXXXXXXXXXXXXXRCNVYSFCKTLTASDTS 617
+NV L+AE D+DEVYAQI L P E D+ R V+SF K LTASDTS
Sbjct: 69 MNVSLQAEKDTDEVYAQITLIPVGTEVDE--PMSPDPSPPELQRPKVHSFSKVLTASDTS 126
Query: 618 THGGFSVLRRHAEE 659
THGGFSVLR+HA E
Sbjct: 127 THGGFSVLRKHATE 140
>sptr|Q9ZTR9|Q9ZTR9 Auxin response factor 8.
Length = 811
Score = 132 bits (331), Expect = 5e-30
Identities = 67/130 (51%), Positives = 91/130 (70%), Gaps = 3/130 (2%)
Frame = +3
Query: 279 ELWHACAGPLVTVPRQGELVYYFPQGHMEQLEASTDQQLDQHLPLF-DLPPKILCKVVNV 455
ELWHACAGPLV++P G V YFPQGH EQ+ A+T++++D H+P + LPP+++C++ NV
Sbjct: 22 ELWHACAGPLVSLPSSGSRVVYFPQGHSEQVAATTNKEVDGHIPNYPSLPPQLICQLHNV 81
Query: 456 ELRAETDSDEVYAQIMLQP--EADQXXXXXXXXXXXXXXRCNVYSFCKTLTASDTSTHGG 629
+ A+ ++DEVYAQ+ LQP +Q + + Y FCKTLTASDTSTHGG
Sbjct: 82 TMHADVETDEVYAQMTLQPLTPEEQKETFVPIELGIPSKQPSNY-FCKTLTASDTSTHGG 140
Query: 630 FSVLRRHAEE 659
FSV RR AE+
Sbjct: 141 FSVPRRAAEK 150
>sptr|Q8S979|Q8S979 Auxin response factor 7a (Fragment).
Length = 1123
Score = 132 bits (331), Expect = 5e-30
Identities = 69/133 (51%), Positives = 86/133 (64%), Gaps = 2/133 (1%)
Frame = +3
Query: 267 ALFRELWHACAGPLVTVPRQGELVYYFPQGHMEQLEASTDQQLDQHLPLF-DLPPKILCK 443
A+ ELWHACAGPLV++P G LV YFPQGH EQ+ AS + +D H+P + +LP K++C
Sbjct: 8 AINSELWHACAGPLVSLPPAGSLVVYFPQGHSEQVAASMQKDVDAHVPSYPNLPSKLICL 67
Query: 444 VVNVELRAETDSDEVYAQIMLQPEADQ-XXXXXXXXXXXXXXRCNVYSFCKTLTASDTST 620
+ NV L A+ ++DEVYAQ+ LQP R FCKTLTASDTST
Sbjct: 68 LHNVTLHADPETDEVYAQMTLQPVTSYGKEALQLSELALKQARPQTEFFCKTLTASDTST 127
Query: 621 HGGFSVLRRHAEE 659
HGGFSV RR AE+
Sbjct: 128 HGGFSVPRRAAEK 140
>sptr|Q9FGV1|Q9FGV1 Auxin responsive transcription factor.
Length = 821
Score = 132 bits (331), Expect = 5e-30
Identities = 67/130 (51%), Positives = 91/130 (70%), Gaps = 3/130 (2%)
Frame = +3
Query: 279 ELWHACAGPLVTVPRQGELVYYFPQGHMEQLEASTDQQLDQHLPLF-DLPPKILCKVVNV 455
ELWHACAGPLV++P G V YFPQGH EQ+ A+T++++D H+P + LPP+++C++ NV
Sbjct: 22 ELWHACAGPLVSLPSSGSRVVYFPQGHSEQVAATTNKEVDGHIPNYPSLPPQLICQLHNV 81
Query: 456 ELRAETDSDEVYAQIMLQP--EADQXXXXXXXXXXXXXXRCNVYSFCKTLTASDTSTHGG 629
+ A+ ++DEVYAQ+ LQP +Q + + Y FCKTLTASDTSTHGG
Sbjct: 82 TMHADVETDEVYAQMTLQPLTPEEQKETFVPIELGIPSKQPSNY-FCKTLTASDTSTHGG 140
Query: 630 FSVLRRHAEE 659
FSV RR AE+
Sbjct: 141 FSVPRRAAEK 150
>sptr|Q9ZTX8|Q9ZTX8 ARF6 (Auxin response factor 6).
Length = 933
Score = 131 bits (329), Expect = 9e-30
Identities = 67/129 (51%), Positives = 88/129 (68%), Gaps = 2/129 (1%)
Frame = +3
Query: 279 ELWHACAGPLVTVPRQGELVYYFPQGHMEQLEASTDQQLDQHLPLF-DLPPKILCKVVNV 455
ELWHACAGPLV++P G V YFPQGH EQ+ AST++++D H+P + L P+++C++ NV
Sbjct: 23 ELWHACAGPLVSLPPVGSRVVYFPQGHSEQVAASTNKEVDAHIPNYPSLHPQLICQLHNV 82
Query: 456 ELRAETDSDEVYAQIMLQP-EADQXXXXXXXXXXXXXXRCNVYSFCKTLTASDTSTHGGF 632
+ A+ ++DEVYAQ+ LQP A + R FCKTLTASDTSTHGGF
Sbjct: 83 TMHADVETDEVYAQMTLQPLNAQEQKDPYLPAELGVPSRQPTNYFCKTLTASDTSTHGGF 142
Query: 633 SVLRRHAEE 659
SV RR AE+
Sbjct: 143 SVPRRAAEK 151
>sptr|Q8S981|Q8S981 Auxin response factor 6a (Fragment).
Length = 396
Score = 130 bits (327), Expect = 1e-29
Identities = 65/129 (50%), Positives = 89/129 (68%), Gaps = 2/129 (1%)
Frame = +3
Query: 279 ELWHACAGPLVTVPRQGELVYYFPQGHMEQLEASTDQQLDQHLPLF-DLPPKILCKVVNV 455
ELWHACAGPLV++P G V YFPQGH EQ+ AST+++++ +P + +LPP+++C++ NV
Sbjct: 9 ELWHACAGPLVSLPAVGSRVVYFPQGHSEQVAASTNKEMESQIPNYPNLPPQLICQLHNV 68
Query: 456 ELRAETDSDEVYAQIMLQPEADQXXXX-XXXXXXXXXXRCNVYSFCKTLTASDTSTHGGF 632
+ A+ ++DEVYAQ+ LQP + Q + FCKTLTASDTSTHGGF
Sbjct: 69 TMHADAETDEVYAQMTLQPLSPQELKDPFLPAELGTASKQPTNYFCKTLTASDTSTHGGF 128
Query: 633 SVLRRHAEE 659
SV RR AE+
Sbjct: 129 SVPRRAAEK 137
>sptr|Q7XM11|Q7XM11 OSJNBa0084K01.20 protein.
Length = 954
Score = 128 bits (322), Expect = 6e-29
Identities = 66/129 (51%), Positives = 87/129 (67%), Gaps = 2/129 (1%)
Frame = +3
Query: 279 ELWHACAGPLVTVPRQGELVYYFPQGHMEQLEASTDQQLDQHLPLF-DLPPKILCKVVNV 455
ELWHACAGPLV +P++G LVYYFPQGH EQ+ A+T + + +P + +LP ++LC+V N+
Sbjct: 39 ELWHACAGPLVCLPQRGSLVYYFPQGHSEQVAATTRKIPNSRIPNYPNLPSQLLCQVHNI 98
Query: 456 ELRAETDSDEVYAQIMLQP-EADQXXXXXXXXXXXXXXRCNVYSFCKTLTASDTSTHGGF 632
L A+ D+DEVYAQ+ LQP ++ + FCK LTASDTSTHGGF
Sbjct: 99 TLHADKDTDEVYAQMTLQPVNSETDVFPIPTLGAYTKSKHPTEYFCKNLTASDTSTHGGF 158
Query: 633 SVLRRHAEE 659
SV RR AE+
Sbjct: 159 SVPRRAAEK 167
>sptr|Q8S983|Q8S983 Arabidopsis Monopteros-like protein.
Length = 955
Score = 128 bits (322), Expect = 6e-29
Identities = 66/129 (51%), Positives = 87/129 (67%), Gaps = 2/129 (1%)
Frame = +3
Query: 279 ELWHACAGPLVTVPRQGELVYYFPQGHMEQLEASTDQQLDQHLPLF-DLPPKILCKVVNV 455
ELWHACAGPLV +P++G LVYYFPQGH EQ+ A+T + + +P + +LP ++LC+V N+
Sbjct: 39 ELWHACAGPLVCLPQRGSLVYYFPQGHSEQVAATTRKIPNSRIPNYPNLPSQLLCQVHNI 98
Query: 456 ELRAETDSDEVYAQIMLQP-EADQXXXXXXXXXXXXXXRCNVYSFCKTLTASDTSTHGGF 632
L A+ D+DEVYAQ+ LQP ++ + FCK LTASDTSTHGGF
Sbjct: 99 TLHADKDTDEVYAQMTLQPVNSETDVFPIPTLGAYTKSKHPTEYFCKNLTASDTSTHGGF 158
Query: 633 SVLRRHAEE 659
SV RR AE+
Sbjct: 159 SVPRRAAEK 167
>sptr|Q8S980|Q8S980 Auxin response factor 6b (Fragment).
Length = 880
Score = 127 bits (318), Expect = 2e-28
Identities = 64/129 (49%), Positives = 88/129 (68%), Gaps = 2/129 (1%)
Frame = +3
Query: 279 ELWHACAGPLVTVPRQGELVYYFPQGHMEQLEASTDQQLDQHLPLF-DLPPKILCKVVNV 455
ELWHACAGPLV++P V YFPQGH EQ+ AST++++D +P + +LPP+++C++ NV
Sbjct: 9 ELWHACAGPLVSLPVVRSRVVYFPQGHSEQVAASTNKEVDAQIPNYPNLPPQLICQLHNV 68
Query: 456 ELRAETDSDEVYAQIMLQP-EADQXXXXXXXXXXXXXXRCNVYSFCKTLTASDTSTHGGF 632
+ A+ ++DEVYAQ+ LQP ++ + FCKTLTASDTSTHGGF
Sbjct: 69 TMHADAETDEVYAQMTLQPLSPEEQKEPFLPMELGAASKQPTNYFCKTLTASDTSTHGGF 128
Query: 633 SVLRRHAEE 659
SV RR AE+
Sbjct: 129 SVPRRAAEK 137
>sptr|Q8GT89|Q8GT89 Hypothetical transcription factor.
Length = 954
Score = 126 bits (317), Expect = 2e-28
Identities = 65/132 (49%), Positives = 85/132 (64%), Gaps = 1/132 (0%)
Frame = +3
Query: 267 ALFRELWHACAGPLVTVPRQGELVYYFPQGHMEQLEASTDQQLDQHLPLF-DLPPKILCK 443
A+ ELWHACAGPLV +P+ G L YYFPQGH EQ+ ST + +P + +LP ++LC+
Sbjct: 42 AINSELWHACAGPLVCLPQVGSLSYYFPQGHSEQVAVSTKRTATSQIPNYPNLPSQLLCQ 101
Query: 444 VVNVELRAETDSDEVYAQIMLQPEADQXXXXXXXXXXXXXXRCNVYSFCKTLTASDTSTH 623
V NV L A+ ++DE+YAQ+ L+P + + FCKTLTASDTSTH
Sbjct: 102 VQNVTLHADKETDEIYAQMSLKPVNSEKDVFPVPDFGLKPSKHPSEFFCKTLTASDTSTH 161
Query: 624 GGFSVLRRHAEE 659
GGFSV RR AE+
Sbjct: 162 GGFSVPRRAAEK 173
>sptr|Q9ZTX9|Q9ZTX9 Auxin response factor 4 (Auxin response factor
ARF4).
Length = 788
Score = 126 bits (317), Expect = 2e-28
Identities = 70/181 (38%), Positives = 94/181 (51%), Gaps = 11/181 (6%)
Frame = +3
Query: 150 VGMG*PDRCSPAEFCSRLVSEAAEMXXXXXXXXXXXXXDALFRELWHACAGPLVTVPRQG 329
VG+G R S ++ +++ ELWHACAGPL +P++G
Sbjct: 22 VGVGGGTRIDKGRLGISPSSSSSCSSGSSSSSSSTGSASSIYSELWHACAGPLTCLPKKG 81
Query: 330 ELVYYFPQGHMEQLEASTDQQLDQHLPLFDLPPKILCKVVNVELRAETDSDEVYAQIMLQ 509
+V YFPQGH+EQ +A +P FDL P+I+C+VVNV+L A D+DEVY Q+ L
Sbjct: 82 NVVVYFPQGHLEQ-DAMVSYSSPLEIPKFDLNPQIVCRVVNVQLLANKDTDEVYTQVTLL 140
Query: 510 PEAD-----------QXXXXXXXXXXXXXXRCNVYSFCKTLTASDTSTHGGFSVLRRHAE 656
P + + + + FCKTLTASDTSTHGGFSV RR AE
Sbjct: 141 PLQEFSMLNGEGKEVKELGGEEERNGSSSVKRTPHMFCKTLTASDTSTHGGFSVPRRAAE 200
Query: 657 E 659
+
Sbjct: 201 D 201
>sptrnew|AAP68244|AAP68244 At1g19850.
Length = 902
Score = 126 bits (316), Expect = 3e-28
Identities = 66/129 (51%), Positives = 85/129 (65%), Gaps = 2/129 (1%)
Frame = +3
Query: 279 ELWHACAGPLVTVPRQGELVYYFPQGHMEQLEASTDQQLDQHLPLF-DLPPKILCKVVNV 455
ELWHACAGPLV +P+ G LVYYF QGH EQ+ ST + +P + +LP +++C+V NV
Sbjct: 54 ELWHACAGPLVCLPQVGSLVYYFSQGHSEQVAVSTRRSATTQVPNYPNLPSQLMCQVHNV 113
Query: 456 ELRAETDSDEVYAQIMLQP-EADQXXXXXXXXXXXXXXRCNVYSFCKTLTASDTSTHGGF 632
L A+ DSDE+YAQ+ LQP +++ + FCKTLTASDTSTHGGF
Sbjct: 114 TLHADKDSDEIYAQMSLQPVHSERDVFPVPDFGMLRGSKHPTEFFCKTLTASDTSTHGGF 173
Query: 633 SVLRRHAEE 659
SV RR AE+
Sbjct: 174 SVPRRAAEK 182
Database: /db/trembl-ebi/tmp/swall
Posted date: Nov 7, 2003 8:22 PM
Number of letters in database: 415,658,970
Number of sequences in database: 1,302,759
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 427,487,921
Number of Sequences: 1302759
Number of extensions: 7476457
Number of successful extensions: 26879
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 25579
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26703
length of database: 415,658,970
effective HSP length: 118
effective length of database: 261,933,408
effective search space used: 26455274208
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

