BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= 2550132.2.1
(603 letters)
Database: /db/uniprot/tmp/swall
1,395,590 sequences; 442,889,342 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
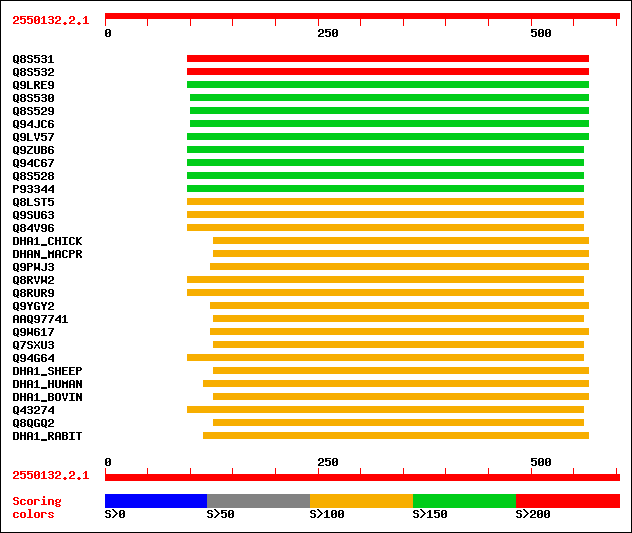
sptr|Q8S531|Q8S531 Cytosolic aldehyde dehydrogenase RF2C. 204 5e-52
sptr|Q8S532|Q8S532 Cytosolic aldehyde dehydrogenase RF2C. 203 2e-51
sptr|Q9LRE9|Q9LRE9 Cytosolic aldehyde dehydrogenase. 197 1e-49
sptr|Q8S530|Q8S530 Cytosolic aldehyde dehydrogenase RF2D (Fragme... 192 4e-48
sptr|Q8S529|Q8S529 Cytosolic aldehyde dehydrogenase RF2D. 192 4e-48
sptr|Q94JC6|Q94JC6 Putative cytosolic aldehyde dehydrogenase. 191 8e-48
sptr|Q9LV57|Q9LV57 Aldehyde dehydrogenase ALDH1a. 175 3e-43
sptr|Q9ZUB6|Q9ZUB6 F5O8.35 protein. 154 8e-37
sptr|Q94C67|Q94C67 Putative aldehyde dehydrogenase NAD+. 154 8e-37
sptr|Q8S528|Q8S528 Mitochondrial aldehyde dehydrogenase. 154 8e-37
sptr|P93344|P93344 Aldehyde dehydrogenase (NAD+) (EC 1.2.1.3). 151 7e-36
sptr|Q8LST5|Q8LST5 Mitochondrial aldehyde dehydrogenase. 147 1e-34
sptr|Q9SU63|Q9SU63 Aldehyde dehydrogenase (NAD+)-like protein (E... 147 1e-34
sptr|Q84V96|Q84V96 Aldehyde dehydrogenase 1 precursor (EC 1.2.1.3). 146 2e-34
sw|P27463|DHA1_CHICK Aldehyde dehydrogenase 1A1 (EC 1.2.1.3) (Al... 145 3e-34
sw|Q29491|DHAN_MACPR Aldehyde dehydrogenase, cytosolic 2 (EC 1.2... 145 5e-34
sptr|Q9PWJ3|Q9PWJ3 Aldehyde dehydrogenase class 1 (EC 1.2.1.3). 144 7e-34
sptr|Q8RVW2|Q8RVW2 Aldehyde dehydrogenase (Fragment). 143 1e-33
sptr|Q8RUR9|Q8RUR9 Mitochondrial aldehyde dehydrogenase RF2B. 143 1e-33
sptr|Q9YGY2|Q9YGY2 Aldehyde dehydrogenase (EC 1.2.1.3). 143 2e-33
sptrnew|AAQ97741|AAQ97741 Mitochondrial aldehyde dehydrogenase 2... 142 2e-33
sptr|Q9W617|Q9W617 Aldehyde dehydrogenase class 1 (EC 1.2.1.3). 142 2e-33
sptr|Q7SXU3|Q7SXU3 Aldehyde dehydrogenase 2. 142 3e-33
sptr|Q94G64|Q94G64 T-cytoplasm male sterility restorer factor 2. 142 4e-33
sw|P51977|DHA1_SHEEP Aldehyde dehydrogenase 1A1 (EC 1.2.1.3) (Al... 142 4e-33
sw|P00352|DHA1_HUMAN Aldehyde dehydrogenase 1A1 (EC 1.2.1.3) (Al... 142 4e-33
sw|P48644|DHA1_BOVIN Aldehyde dehydrogenase 1A1 (EC 1.2.1.3) (Al... 142 4e-33
sptr|Q43274|Q43274 T CYTOPLASM MALE sterility RESTORER factor 2 ... 141 6e-33
sptr|Q8QGQ2|Q8QGQ2 Aldehyde dehydrogenase 2. 141 6e-33
sw|Q8MI17|DHA1_RABIT Aldehyde dehydrogenase 1A1 (EC 1.2.1.3) (Al... 141 7e-33
>sptr|Q8S531|Q8S531 Cytosolic aldehyde dehydrogenase RF2C.
Length = 503
Score = 204 bits (520), Expect = 5e-52
Identities = 100/157 (63%), Positives = 118/157 (75%)
Frame = -1
Query: 567 KAQYERVLSYIGHGKREGATLLTGGRPCGPEGKGYYIEPTVFTNVKEDMIIAKEEIFGPV 388
K QYE+VL YI GKREGATL+TGG+PCG + KGYYIEPT+FT+VK+DM IA++EIFGPV
Sbjct: 348 KDQYEKVLRYIDIGKREGATLVTGGKPCG-DNKGYYIEPTIFTDVKDDMTIAQDEIFGPV 406
Query: 387 MCLMKFKPVEEAIARANDTRYGLGAGVVTRDLDXXXXXXXXXXXXXXXXNCYFAMGSDCP 208
M LMKFK VEE I +AN+TRYGL AG+VT+++D NCYFA D P
Sbjct: 407 MALMKFKTVEEVIQKANNTRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAP 466
Query: 207 FGGRKMSGFGKDEGMHALDKYLAVKSVVTPLRASPWI 97
FGG KMSGFGKD GM ALDKYL K+VVTPL +PW+
Sbjct: 467 FGGYKMSGFGKDMGMDALDKYLQTKTVVTPLYNTPWL 503
>sptr|Q8S532|Q8S532 Cytosolic aldehyde dehydrogenase RF2C.
Length = 502
Score = 203 bits (516), Expect = 2e-51
Identities = 100/157 (63%), Positives = 117/157 (74%)
Frame = -1
Query: 567 KAQYERVLSYIGHGKREGATLLTGGRPCGPEGKGYYIEPTVFTNVKEDMIIAKEEIFGPV 388
K QYE+VL YI GKREGATL+TGG+PCG KGYYIEPT+FT+VK+DM IA++EIFGPV
Sbjct: 348 KDQYEKVLRYIDIGKREGATLVTGGKPCGD--KGYYIEPTIFTDVKDDMTIAQDEIFGPV 405
Query: 387 MCLMKFKPVEEAIARANDTRYGLGAGVVTRDLDXXXXXXXXXXXXXXXXNCYFAMGSDCP 208
M LMKFK VEE I +AN+TRYGL AG+VT+++D NCYFA D P
Sbjct: 406 MALMKFKTVEEVIQKANNTRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAP 465
Query: 207 FGGRKMSGFGKDEGMHALDKYLAVKSVVTPLRASPWI 97
FGG KMSGFGKD GM ALDKYL K+VVTPL +PW+
Sbjct: 466 FGGYKMSGFGKDMGMDALDKYLQTKTVVTPLYNTPWL 502
>sptr|Q9LRE9|Q9LRE9 Cytosolic aldehyde dehydrogenase.
Length = 502
Score = 197 bits (500), Expect = 1e-49
Identities = 97/157 (61%), Positives = 114/157 (72%)
Frame = -1
Query: 567 KAQYERVLSYIGHGKREGATLLTGGRPCGPEGKGYYIEPTVFTNVKEDMIIAKEEIFGPV 388
K QYE++L YI GKREGATL+TGG+PCG G YYIEPT+FT+VKE+M IA+EEIFGPV
Sbjct: 348 KEQYEKILKYIDIGKREGATLVTGGKPCGENG--YYIEPTIFTDVKEEMSIAQEEIFGPV 405
Query: 387 MCLMKFKPVEEAIARANDTRYGLGAGVVTRDLDXXXXXXXXXXXXXXXXNCYFAMGSDCP 208
M LMKFK VEEAI +AN TRYGL AG+VT+++D NCY D P
Sbjct: 406 MALMKFKTVEEAIQKANSTRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYLGFDPDVP 465
Query: 207 FGGRKMSGFGKDEGMHALDKYLAVKSVVTPLRASPWI 97
FGG KMSGFGKD GM AL+KYL K+VVTPL +PW+
Sbjct: 466 FGGYKMSGFGKDMGMDALEKYLHTKAVVTPLYNTPWL 502
>sptr|Q8S530|Q8S530 Cytosolic aldehyde dehydrogenase RF2D
(Fragment).
Length = 466
Score = 192 bits (487), Expect = 4e-48
Identities = 97/156 (62%), Positives = 111/156 (71%)
Frame = -1
Query: 567 KAQYERVLSYIGHGKREGATLLTGGRPCGPEGKGYYIEPTVFTNVKEDMIIAKEEIFGPV 388
K Q+ERVL YI HGK EGATLLTGG+P KGYYIEPT+F +V EDM IA+EEIFGPV
Sbjct: 312 KDQFERVLKYIEHGKSEGATLLTGGKPAAD--KGYYIEPTIFVDVTEDMKIAQEEIFGPV 369
Query: 387 MCLMKFKPVEEAIARANDTRYGLGAGVVTRDLDXXXXXXXXXXXXXXXXNCYFAMGSDCP 208
M LMKFK V+E I +AN TRYGL AG+VT+ LD NCYFA D P
Sbjct: 370 MSLMKFKTVDEVIEKANCTRYGLAAGIVTKSLDVANRVSRSVRAGTVWVNCYFAFDPDAP 429
Query: 207 FGGRKMSGFGKDEGMHALDKYLAVKSVVTPLRASPW 100
FGG KMSGFG+D+G+ A+DKYL VKSV+T L SPW
Sbjct: 430 FGGYKMSGFGRDQGLAAMDKYLQVKSVITALPDSPW 465
>sptr|Q8S529|Q8S529 Cytosolic aldehyde dehydrogenase RF2D.
Length = 511
Score = 192 bits (487), Expect = 4e-48
Identities = 97/156 (62%), Positives = 111/156 (71%)
Frame = -1
Query: 567 KAQYERVLSYIGHGKREGATLLTGGRPCGPEGKGYYIEPTVFTNVKEDMIIAKEEIFGPV 388
K Q+ERVL YI HGK EGATLLTGG+P KGYYIEPT+F +V EDM IA+EEIFGPV
Sbjct: 357 KDQFERVLKYIEHGKSEGATLLTGGKPAAD--KGYYIEPTIFVDVTEDMKIAQEEIFGPV 414
Query: 387 MCLMKFKPVEEAIARANDTRYGLGAGVVTRDLDXXXXXXXXXXXXXXXXNCYFAMGSDCP 208
M LMKFK V+E I +AN TRYGL AG+VT+ LD NCYFA D P
Sbjct: 415 MSLMKFKTVDEVIEKANCTRYGLAAGIVTKSLDVANRVSRSVRAGTVWVNCYFAFDPDAP 474
Query: 207 FGGRKMSGFGKDEGMHALDKYLAVKSVVTPLRASPW 100
FGG KMSGFG+D+G+ A+DKYL VKSV+T L SPW
Sbjct: 475 FGGYKMSGFGRDQGLAAMDKYLQVKSVITALPDSPW 510
>sptr|Q94JC6|Q94JC6 Putative cytosolic aldehyde dehydrogenase.
Length = 507
Score = 191 bits (484), Expect = 8e-48
Identities = 96/156 (61%), Positives = 114/156 (73%)
Frame = -1
Query: 567 KAQYERVLSYIGHGKREGATLLTGGRPCGPEGKGYYIEPTVFTNVKEDMIIAKEEIFGPV 388
K Q+ERVL YI GK EGATLLTGG+P G KGYYIEPT+F +VKE+M IA+EEIFGPV
Sbjct: 353 KVQFERVLKYIEIGKNEGATLLTGGKPTGD--KGYYIEPTIFVDVKEEMTIAQEEIFGPV 410
Query: 387 MCLMKFKPVEEAIARANDTRYGLGAGVVTRDLDXXXXXXXXXXXXXXXXNCYFAMGSDCP 208
M LMKFK VEEAI +AN T+YGL AG+VT++L+ NCYFA D P
Sbjct: 411 MSLMKFKTVEEAIEKANCTKYGLAAGIVTKNLNIANMVSRSVRAGTVWVNCYFAFDPDAP 470
Query: 207 FGGRKMSGFGKDEGMHALDKYLAVKSVVTPLRASPW 100
FGG KMSGFG+D+GM A+DKYL VK+V+T + SPW
Sbjct: 471 FGGYKMSGFGRDQGMVAMDKYLQVKTVITAVPDSPW 506
>sptr|Q9LV57|Q9LV57 Aldehyde dehydrogenase ALDH1a.
Length = 501
Score = 175 bits (444), Expect = 3e-43
Identities = 87/157 (55%), Positives = 108/157 (68%)
Frame = -1
Query: 567 KAQYERVLSYIGHGKREGATLLTGGRPCGPEGKGYYIEPTVFTNVKEDMIIAKEEIFGPV 388
K Q+E++LSYI HGK EGATLLTGG+ G KGY+I+PT+F +V EDM I ++EIFGPV
Sbjct: 347 KRQFEKILSYIEHGKNEGATLLTGGKAIGD--KGYFIQPTIFADVTEDMKIYQDEIFGPV 404
Query: 387 MCLMKFKPVEEAIARANDTRYGLGAGVVTRDLDXXXXXXXXXXXXXXXXNCYFAMGSDCP 208
M LMKFK VEE I AN+T+YGL AG++++D+D NCYF DCP
Sbjct: 405 MSLMKFKTVEEGIKCANNTKYGLAAGILSQDIDLINTVSRSIKAGIIWVNCYFGFDLDCP 464
Query: 207 FGGRKMSGFGKDEGMHALDKYLAVKSVVTPLRASPWI 97
+GG KMSG ++ GM ALD YL KSVV PL SPW+
Sbjct: 465 YGGYKMSGNCRESGMDALDNYLQTKSVVMPLHNSPWM 501
>sptr|Q9ZUB6|Q9ZUB6 F5O8.35 protein.
Length = 519
Score = 154 bits (389), Expect = 8e-37
Identities = 75/155 (48%), Positives = 105/155 (67%)
Frame = -1
Query: 561 QYERVLSYIGHGKREGATLLTGGRPCGPEGKGYYIEPTVFTNVKEDMIIAKEEIFGPVMC 382
Q+ ++L YI HG GATL GG G KGYYI+PTVF++VK+DM+IA +EIFGPV
Sbjct: 367 QFNKILKYIKHGVEAGATLQAGGDRLG--SKGYYIQPTVFSDVKDDMLIATDEIFGPVQT 424
Query: 381 LMKFKPVEEAIARANDTRYGLGAGVVTRDLDXXXXXXXXXXXXXXXXNCYFAMGSDCPFG 202
++KFK ++E IARAN++RYGL AGV T++LD NC+ + + PFG
Sbjct: 425 ILKFKDLDEVIARANNSRYGLAAGVFTQNLDTAHRLMRALRVGTVWINCFDVLDASIPFG 484
Query: 201 GRKMSGFGKDEGMHALDKYLAVKSVVTPLRASPWI 97
G KMSG G+++G+++L+ YL VK+VVT L+ W+
Sbjct: 485 GYKMSGIGREKGIYSLNNYLQVKAVVTSLKNPAWL 519
>sptr|Q94C67|Q94C67 Putative aldehyde dehydrogenase NAD+.
Length = 534
Score = 154 bits (389), Expect = 8e-37
Identities = 75/155 (48%), Positives = 105/155 (67%)
Frame = -1
Query: 561 QYERVLSYIGHGKREGATLLTGGRPCGPEGKGYYIEPTVFTNVKEDMIIAKEEIFGPVMC 382
Q+ ++L YI HG GATL GG G KGYYI+PTVF++VK+DM+IA +EIFGPV
Sbjct: 382 QFNKILKYIKHGVEAGATLQAGGDRLG--SKGYYIQPTVFSDVKDDMLIATDEIFGPVQT 439
Query: 381 LMKFKPVEEAIARANDTRYGLGAGVVTRDLDXXXXXXXXXXXXXXXXNCYFAMGSDCPFG 202
++KFK ++E IARAN++RYGL AGV T++LD NC+ + + PFG
Sbjct: 440 ILKFKDLDEVIARANNSRYGLAAGVFTQNLDTAHRLMRALRVGTVWINCFDVLDASIPFG 499
Query: 201 GRKMSGFGKDEGMHALDKYLAVKSVVTPLRASPWI 97
G KMSG G+++G+++L+ YL VK+VVT L+ W+
Sbjct: 500 GYKMSGIGREKGIYSLNNYLQVKAVVTSLKNPAWL 534
>sptr|Q8S528|Q8S528 Mitochondrial aldehyde dehydrogenase.
Length = 534
Score = 154 bits (389), Expect = 8e-37
Identities = 75/155 (48%), Positives = 105/155 (67%)
Frame = -1
Query: 561 QYERVLSYIGHGKREGATLLTGGRPCGPEGKGYYIEPTVFTNVKEDMIIAKEEIFGPVMC 382
Q+ ++L YI HG GATL GG G KGYYI+PTVF++VK+DM+IA +EIFGPV
Sbjct: 382 QFNKILKYIKHGVEAGATLQAGGDRLG--SKGYYIQPTVFSDVKDDMLIATDEIFGPVQT 439
Query: 381 LMKFKPVEEAIARANDTRYGLGAGVVTRDLDXXXXXXXXXXXXXXXXNCYFAMGSDCPFG 202
++KFK ++E IARAN++RYGL AGV T++LD NC+ + + PFG
Sbjct: 440 ILKFKDLDEVIARANNSRYGLAAGVFTQNLDTAHRLMRALRVGTVWINCFDVLDASIPFG 499
Query: 201 GRKMSGFGKDEGMHALDKYLAVKSVVTPLRASPWI 97
G KMSG G+++G+++L+ YL VK+VVT L+ W+
Sbjct: 500 GYKMSGIGREKGIYSLNNYLQVKAVVTSLKNPAWL 534
>sptr|P93344|P93344 Aldehyde dehydrogenase (NAD+) (EC 1.2.1.3).
Length = 542
Score = 151 bits (381), Expect = 7e-36
Identities = 73/155 (47%), Positives = 105/155 (67%)
Frame = -1
Query: 561 QYERVLSYIGHGKREGATLLTGGRPCGPEGKGYYIEPTVFTNVKEDMIIAKEEIFGPVMC 382
Q++++++YI G GATL TGG G +GYYI+PTVF+NVK+DM+IA++EIFGPV
Sbjct: 390 QFDKIMNYIRSGIDSGATLETGGERLGE--RGYYIKPTVFSNVKDDMLIAQDEIFGPVQS 447
Query: 381 LMKFKPVEEAIARANDTRYGLGAGVVTRDLDXXXXXXXXXXXXXXXXNCYFAMGSDCPFG 202
++KFK V++ I RAN++RYGL AGV T+++D NC+ + PFG
Sbjct: 448 ILKFKDVDDVIRRANNSRYGLAAGVFTQNIDTANTLTRALRVGTVWVNCFDTFDATIPFG 507
Query: 201 GRKMSGFGKDEGMHALDKYLAVKSVVTPLRASPWI 97
G KMSG G+++G ++L YL VK+VVTPL+ W+
Sbjct: 508 GYKMSGHGREKGEYSLKNYLQVKAVVTPLKNPAWL 542
>sptr|Q8LST5|Q8LST5 Mitochondrial aldehyde dehydrogenase.
Length = 551
Score = 147 bits (370), Expect = 1e-34
Identities = 73/155 (47%), Positives = 97/155 (62%)
Frame = -1
Query: 561 QYERVLSYIGHGKREGATLLTGGRPCGPEGKGYYIEPTVFTNVKEDMIIAKEEIFGPVMC 382
Q+ ++L Y+ G GATL+TGG G +G+YI+PTVF + K+DM IA+EEIFGPV
Sbjct: 399 QFNKILRYVQSGVDSGATLVTGGDRVG--SRGFYIQPTVFADAKDDMKIAREEIFGPVQT 456
Query: 381 LMKFKPVEEAIARANDTRYGLGAGVVTRDLDXXXXXXXXXXXXXXXXNCYFAMGSDCPFG 202
++KF +EE I RAN T YGL AGV TR LD NCY + PFG
Sbjct: 457 ILKFSGMEEVIRRANATHYGLAAGVFTRSLDAANTLSRALRAGTVWVNCYDVFDATIPFG 516
Query: 201 GRKMSGFGKDEGMHALDKYLAVKSVVTPLRASPWI 97
G KMSG G+++G++AL YL K+VVTP++ W+
Sbjct: 517 GYKMSGVGREKGVYALRNYLQTKAVVTPIKDPAWL 551
>sptr|Q9SU63|Q9SU63 Aldehyde dehydrogenase (NAD+)-like protein (EC
1.2.1.3) (Putative aldehyde dehydrogenase (NAD+)
protein) (AT3g48000/T17F15_130) (Aldehyde dehydrogenase
ALDH2a) (NAD+).
Length = 538
Score = 147 bits (370), Expect = 1e-34
Identities = 73/155 (47%), Positives = 101/155 (65%)
Frame = -1
Query: 561 QYERVLSYIGHGKREGATLLTGGRPCGPEGKGYYIEPTVFTNVKEDMIIAKEEIFGPVMC 382
Q+E+V+ YI G ATL GG G KGY+I+PTVF+NVK+DM+IA++EIFGPV
Sbjct: 386 QFEKVMKYIKSGIESNATLECGGDQIGD--KGYFIQPTVFSNVKDDMLIAQDEIFGPVQS 443
Query: 381 LMKFKPVEEAIARANDTRYGLGAGVVTRDLDXXXXXXXXXXXXXXXXNCYFAMGSDCPFG 202
++KF V+E I RAN+T+YGL AGV T++LD NC+ + PFG
Sbjct: 444 ILKFSDVDEVIKRANETKYGLAAGVFTKNLDTANRVSRALKAGTVWVNCFDVFDAAIPFG 503
Query: 201 GRKMSGFGKDEGMHALDKYLAVKSVVTPLRASPWI 97
G KMSG G+++G+++L+ YL +K+VVT L WI
Sbjct: 504 GYKMSGNGREKGIYSLNNYLQIKAVVTALNKPAWI 538
>sptr|Q84V96|Q84V96 Aldehyde dehydrogenase 1 precursor (EC 1.2.1.3).
Length = 542
Score = 146 bits (368), Expect = 2e-34
Identities = 72/155 (46%), Positives = 100/155 (64%)
Frame = -1
Query: 561 QYERVLSYIGHGKREGATLLTGGRPCGPEGKGYYIEPTVFTNVKEDMIIAKEEIFGPVMC 382
Q+E++L YI G ATL GG G KG+Y++PTVF+NV++DM+IA++EIFGPV
Sbjct: 390 QFEKILRYIKSGIESNATLECGGDRLG--SKGFYVQPTVFSNVQDDMLIAQDEIFGPVQT 447
Query: 381 LMKFKPVEEAIARANDTRYGLGAGVVTRDLDXXXXXXXXXXXXXXXXNCYFAMGSDCPFG 202
+ KFK ++E I RAN TRYGL AGV T++L NC+ + PFG
Sbjct: 448 IFKFKEIDEVIRRANSTRYGLAAGVFTQNLATANTLMRALRAGTVWINCFDVFDAAIPFG 507
Query: 201 GRKMSGFGKDEGMHALDKYLAVKSVVTPLRASPWI 97
G KMSG G+++G+++L YL VK+VVTPL+ W+
Sbjct: 508 GYKMSGIGREKGIYSLHNYLQVKAVVTPLKNPAWL 542
>sw|P27463|DHA1_CHICK Aldehyde dehydrogenase 1A1 (EC 1.2.1.3)
(Aldehyde dehydrogenase, cytosolic) (ALDH class 1).
Length = 509
Score = 145 bits (367), Expect = 3e-34
Identities = 74/147 (50%), Positives = 95/147 (64%)
Frame = -1
Query: 567 KAQYERVLSYIGHGKREGATLLTGGRPCGPEGKGYYIEPTVFTNVKEDMIIAKEEIFGPV 388
K Q++++L I GK+EGA L GG P G KGY+I+PTVF+NV +DM IAKEEIFGPV
Sbjct: 356 KEQFQKILDLIESGKKEGAKLECGGGPWG--NKGYFIQPTVFSNVTDDMRIAKEEIFGPV 413
Query: 387 MCLMKFKPVEEAIARANDTRYGLGAGVVTRDLDXXXXXXXXXXXXXXXXNCYFAMGSDCP 208
+MKFK ++E I RAN+T YGL A V T+D+D NCY A + CP
Sbjct: 414 QQIMKFKTIDEVIKRANNTTYGLAAAVFTKDIDKALTFASALQAGTVWVNCYSAFSAQCP 473
Query: 207 FGGRKMSGFGKDEGMHALDKYLAVKSV 127
FGG KMSG G++ G + L +Y VK+V
Sbjct: 474 FGGFKMSGNGRELGEYGLQEYTEVKTV 500
>sw|Q29491|DHAN_MACPR Aldehyde dehydrogenase, cytosolic 2 (EC
1.2.1.3) (ALDH class 1) (Non- lens ALDH1) (ALDH1-NL)
(Fragment).
Length = 240
Score = 145 bits (365), Expect = 5e-34
Identities = 75/147 (51%), Positives = 95/147 (64%)
Frame = -1
Query: 567 KAQYERVLSYIGHGKREGATLLTGGRPCGPEGKGYYIEPTVFTNVKEDMIIAKEEIFGPV 388
K QY++++ I GK+EGA L GG P G KGY+I+PTVF+NV ++M IAKEEIFGPV
Sbjct: 87 KEQYDKIIDLIESGKKEGAKLECGGGPWG--NKGYFIQPTVFSNVTDEMRIAKEEIFGPV 144
Query: 387 MCLMKFKPVEEAIARANDTRYGLGAGVVTRDLDXXXXXXXXXXXXXXXXNCYFAMGSDCP 208
+MKFK ++E I RAN+T YGL AGV T+DLD NCY A CP
Sbjct: 145 QQIMKFKSLDEVIKRANNTFYGLAAGVFTKDLDKAVTVSAALQAGTVWVNCYMANSVQCP 204
Query: 207 FGGRKMSGFGKDEGMHALDKYLAVKSV 127
FGG KMSG G++ G + L +Y VK+V
Sbjct: 205 FGGFKMSGNGRELGEYGLHEYTEVKTV 231
>sptr|Q9PWJ3|Q9PWJ3 Aldehyde dehydrogenase class 1 (EC 1.2.1.3).
Length = 502
Score = 144 bits (364), Expect = 7e-34
Identities = 74/148 (50%), Positives = 96/148 (64%)
Frame = -1
Query: 567 KAQYERVLSYIGHGKREGATLLTGGRPCGPEGKGYYIEPTVFTNVKEDMIIAKEEIFGPV 388
K QY+++L I GK+EGA L GG G KG+YI PTVF++VK+DM IAKEEIFGPV
Sbjct: 349 KEQYDKILELIESGKKEGAKLQCGGSAWGE--KGFYISPTVFSDVKDDMRIAKEEIFGPV 406
Query: 387 MCLMKFKPVEEAIARANDTRYGLGAGVVTRDLDXXXXXXXXXXXXXXXXNCYFAMGSDCP 208
++KFK ++E I RAN+T+YGL AGV T+D+D NCY AM P
Sbjct: 407 QQILKFKTIDEVIKRANNTKYGLAAGVFTKDMDKAILMSTALQAGTVWINCYSAMSPQSP 466
Query: 207 FGGRKMSGFGKDEGMHALDKYLAVKSVV 124
FGG KMSG G++ G + L +Y VK+V+
Sbjct: 467 FGGFKMSGNGREMGEYGLHEYTEVKTVI 494
>sptr|Q8RVW2|Q8RVW2 Aldehyde dehydrogenase (Fragment).
Length = 230
Score = 143 bits (361), Expect = 1e-33
Identities = 72/155 (46%), Positives = 99/155 (63%)
Frame = -1
Query: 561 QYERVLSYIGHGKREGATLLTGGRPCGPEGKGYYIEPTVFTNVKEDMIIAKEEIFGPVMC 382
Q+ ++L YI G GA+L+TGG G KGYYI+PT+F++VK+DM+IAKEEIFGPV
Sbjct: 78 QFNKILRYIKLGVDGGASLVTGGERIG--SKGYYIQPTIFSDVKDDMVIAKEEIFGPVQS 135
Query: 381 LMKFKPVEEAIARANDTRYGLGAGVVTRDLDXXXXXXXXXXXXXXXXNCYFAMGSDCPFG 202
++KF + E I RAN +RYGL AGV T ++D NC+ + PFG
Sbjct: 136 ILKFNDLNEVIRRANASRYGLAAGVFTSNIDKANTLTRALRVGTVWVNCFDVFDAAIPFG 195
Query: 201 GRKMSGFGKDEGMHALDKYLAVKSVVTPLRASPWI 97
G KMSG G+++G+ +L YL K+VVTPL+ W+
Sbjct: 196 GYKMSGQGREKGIDSLKGYLQTKAVVTPLKNPAWL 230
>sptr|Q8RUR9|Q8RUR9 Mitochondrial aldehyde dehydrogenase RF2B.
Length = 550
Score = 143 bits (361), Expect = 1e-33
Identities = 72/155 (46%), Positives = 96/155 (61%)
Frame = -1
Query: 561 QYERVLSYIGHGKREGATLLTGGRPCGPEGKGYYIEPTVFTNVKEDMIIAKEEIFGPVMC 382
Q+ ++L Y+ G GATL+ GG G +G+YI+PTVF + K++M IA+EEIFGPV
Sbjct: 398 QFNKILRYVQSGVDSGATLVAGGDRVGD--RGFYIQPTVFADAKDEMKIAREEIFGPVQT 455
Query: 381 LMKFKPVEEAIARANDTRYGLGAGVVTRDLDXXXXXXXXXXXXXXXXNCYFAMGSDCPFG 202
++KF VEE I RAN T YGL AGV TR LD NCY + PFG
Sbjct: 456 ILKFSGVEEVIRRANATPYGLAAGVFTRSLDAANTLSRALRAGTVWVNCYDVFDATIPFG 515
Query: 201 GRKMSGFGKDEGMHALDKYLAVKSVVTPLRASPWI 97
G KMSG G+++G++AL YL K+VVTP++ W+
Sbjct: 516 GYKMSGVGREKGIYALRNYLQTKAVVTPIKNPAWL 550
>sptr|Q9YGY2|Q9YGY2 Aldehyde dehydrogenase (EC 1.2.1.3).
Length = 502
Score = 143 bits (360), Expect = 2e-33
Identities = 74/148 (50%), Positives = 95/148 (64%)
Frame = -1
Query: 567 KAQYERVLSYIGHGKREGATLLTGGRPCGPEGKGYYIEPTVFTNVKEDMIIAKEEIFGPV 388
K QY++ L I GK+EGA L GG G KG+YI PTVF++VK+DM IAKEEIFGPV
Sbjct: 349 KEQYDKCLELIESGKKEGAKLQCGGSAWGE--KGFYISPTVFSDVKDDMRIAKEEIFGPV 406
Query: 387 MCLMKFKPVEEAIARANDTRYGLGAGVVTRDLDXXXXXXXXXXXXXXXXNCYFAMGSDCP 208
++KFK ++E I RAN+T+YGL AGV T+D+D NCY AM P
Sbjct: 407 QQILKFKTIDEVIKRANNTKYGLAAGVFTKDMDKAILMSTALQAGTVWINCYSAMSPQSP 466
Query: 207 FGGRKMSGFGKDEGMHALDKYLAVKSVV 124
FGG KMSG G++ G + L +Y VK+V+
Sbjct: 467 FGGFKMSGNGREMGEYGLHEYTEVKTVI 494
>sptrnew|AAQ97741|AAQ97741 Mitochondrial aldehyde dehydrogenase 2
family.
Length = 516
Score = 142 bits (359), Expect = 2e-33
Identities = 71/145 (48%), Positives = 93/145 (64%)
Frame = -1
Query: 561 QYERVLSYIGHGKREGATLLTGGRPCGPEGKGYYIEPTVFTNVKEDMIIAKEEIFGPVMC 382
Q+++VL YI GKREGA L+ GG P +GY+I+PTVF +VK+DM IA+EEIFGPVM
Sbjct: 365 QFKKVLGYISSGKREGAKLMCGGAPAAE--RGYFIQPTVFGDVKDDMTIAREEIFGPVMQ 422
Query: 381 LMKFKPVEEAIARANDTRYGLGAGVVTRDLDXXXXXXXXXXXXXXXXNCYFAMGSDCPFG 202
++KFK +EE I RAND++YGL V T+D+D NCY G PFG
Sbjct: 423 ILKFKSLEEVIERANDSKYGLAGAVFTQDIDKANYISHGLRAGTVWINCYNVFGVQAPFG 482
Query: 201 GRKMSGFGKDEGMHALDKYLAVKSV 127
G K SG G++ G + L+ Y VK+V
Sbjct: 483 GYKASGIGREMGEYGLENYTEVKTV 507
>sptr|Q9W617|Q9W617 Aldehyde dehydrogenase class 1 (EC 1.2.1.3).
Length = 414
Score = 142 bits (359), Expect = 2e-33
Identities = 74/148 (50%), Positives = 94/148 (63%)
Frame = -1
Query: 567 KAQYERVLSYIGHGKREGATLLTGGRPCGPEGKGYYIEPTVFTNVKEDMIIAKEEIFGPV 388
K QY ++L I GK+EGA L GG G KG+YI PTVF++VK+DM IAKEEIFGPV
Sbjct: 261 KEQYNKILELIESGKKEGAKLECGGSAWGE--KGFYISPTVFSDVKDDMRIAKEEIFGPV 318
Query: 387 MCLMKFKPVEEAIARANDTRYGLGAGVVTRDLDXXXXXXXXXXXXXXXXNCYFAMGSDCP 208
++KFK ++E I RAN+T YGL AGV T+D+D NCY AM P
Sbjct: 319 QQILKFKTIDEVIKRANNTNYGLAAGVFTKDMDKAILMSTALQAGTVWINCYSAMSPQSP 378
Query: 207 FGGRKMSGFGKDEGMHALDKYLAVKSVV 124
FGG KMSG G++ G + L +Y VK+V+
Sbjct: 379 FGGFKMSGNGREMGEYGLHEYTEVKTVI 406
>sptr|Q7SXU3|Q7SXU3 Aldehyde dehydrogenase 2.
Length = 516
Score = 142 bits (358), Expect = 3e-33
Identities = 72/145 (49%), Positives = 94/145 (64%)
Frame = -1
Query: 561 QYERVLSYIGHGKREGATLLTGGRPCGPEGKGYYIEPTVFTNVKEDMIIAKEEIFGPVMC 382
Q+++VL YI GKREGA L+ GG P +GY+I+PTVF +VK+DM IA+EEIFGPVM
Sbjct: 365 QFKKVLGYISSGKREGAKLMCGGAPAAE--RGYFIQPTVFGDVKDDMTIAREEIFGPVMQ 422
Query: 381 LMKFKPVEEAIARANDTRYGLGAGVVTRDLDXXXXXXXXXXXXXXXXNCYFAMGSDCPFG 202
++KFK +EE I RAND++YGL A V T+++D NCY G PFG
Sbjct: 423 ILKFKSLEEVIERANDSKYGLAAAVFTQNIDKANYISHGLRAGTVWINCYNVFGVQAPFG 482
Query: 201 GRKMSGFGKDEGMHALDKYLAVKSV 127
G K SG G++ G + LD Y VK+V
Sbjct: 483 GYKASGIGRELGEYGLDIYTEVKTV 507
>sptr|Q94G64|Q94G64 T-cytoplasm male sterility restorer factor 2.
Length = 549
Score = 142 bits (357), Expect = 4e-33
Identities = 70/155 (45%), Positives = 101/155 (65%)
Frame = -1
Query: 561 QYERVLSYIGHGKREGATLLTGGRPCGPEGKGYYIEPTVFTNVKEDMIIAKEEIFGPVMC 382
Q++++L YI +G GATL+TGG G KG+YI+PT+F++V++ M IA+EEIFGPV
Sbjct: 397 QFKKILRYIRYGVDGGATLVTGGDRLGD--KGFYIQPTIFSDVQDGMKIAQEEIFGPVQS 454
Query: 381 LMKFKPVEEAIARANDTRYGLGAGVVTRDLDXXXXXXXXXXXXXXXXNCYFAMGSDCPFG 202
++KFK + E I RAN ++YGL AGV T LD NC+ + PFG
Sbjct: 455 ILKFKDLNEVIKRANASQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFG 514
Query: 201 GRKMSGFGKDEGMHALDKYLAVKSVVTPLRASPWI 97
G KMSG G+++G+ +L YL VK+VVTP++ + W+
Sbjct: 515 GYKMSGMGREKGVDSLKNYLQVKAVVTPIKNAAWL 549
>sw|P51977|DHA1_SHEEP Aldehyde dehydrogenase 1A1 (EC 1.2.1.3)
(Aldehyde dehydrogenase, cytosolic) (ALDH class 1)
(ALHDII) (ALDH-E1).
Length = 500
Score = 142 bits (357), Expect = 4e-33
Identities = 72/147 (48%), Positives = 94/147 (63%)
Frame = -1
Query: 567 KAQYERVLSYIGHGKREGATLLTGGRPCGPEGKGYYIEPTVFTNVKEDMIIAKEEIFGPV 388
K QYE++L I GK+EGA L GG P G KGY+I+PTVF++V +DM IAKEEIFGPV
Sbjct: 347 KEQYEKILDLIESGKKEGAKLECGGGPWG--NKGYFIQPTVFSDVTDDMRIAKEEIFGPV 404
Query: 387 MCLMKFKPVEEAIARANDTRYGLGAGVVTRDLDXXXXXXXXXXXXXXXXNCYFAMGSDCP 208
+MKFK +++ I RAN+T YGL AG+ T D+D NCY + + CP
Sbjct: 405 QQIMKFKSLDDVIKRANNTFYGLSAGIFTNDIDKAITVSSALQSGTVWVNCYSVVSAQCP 464
Query: 207 FGGRKMSGFGKDEGMHALDKYLAVKSV 127
FGG KMSG G++ G + +Y VK+V
Sbjct: 465 FGGFKMSGNGRELGEYGFHEYTEVKTV 491
>sw|P00352|DHA1_HUMAN Aldehyde dehydrogenase 1A1 (EC 1.2.1.3)
(Aldehyde dehydrogenase, cytosolic) (ALDH class 1)
(Retinal dehydrogenase 1) (ALHDII) (ALDH-E1).
Length = 500
Score = 142 bits (357), Expect = 4e-33
Identities = 71/151 (47%), Positives = 96/151 (63%)
Frame = -1
Query: 567 KAQYERVLSYIGHGKREGATLLTGGRPCGPEGKGYYIEPTVFTNVKEDMIIAKEEIFGPV 388
K QY+++L I GK+EGA L GG P G KGY+++PTVF+NV ++M IAKEEIFGPV
Sbjct: 347 KEQYDKILDLIESGKKEGAKLECGGGPWG--NKGYFVQPTVFSNVTDEMRIAKEEIFGPV 404
Query: 387 MCLMKFKPVEEAIARANDTRYGLGAGVVTRDLDXXXXXXXXXXXXXXXXNCYFAMGSDCP 208
+MKFK +++ I RAN+T YGL AGV T+D+D NCY + + CP
Sbjct: 405 QQIMKFKSLDDVIKRANNTFYGLSAGVFTKDIDKAITISSALQAGTVWVNCYGVVSAQCP 464
Query: 207 FGGRKMSGFGKDEGMHALDKYLAVKSVVTPL 115
FGG KMSG G++ G + +Y VK+V +
Sbjct: 465 FGGFKMSGNGRELGEYGFHEYTEVKTVTVKI 495
>sw|P48644|DHA1_BOVIN Aldehyde dehydrogenase 1A1 (EC 1.2.1.3)
(Aldehyde dehydrogenase, cytosolic) (ALDH class 1)
(ALHDII) (ALDH-E1).
Length = 500
Score = 142 bits (357), Expect = 4e-33
Identities = 72/147 (48%), Positives = 94/147 (63%)
Frame = -1
Query: 567 KAQYERVLSYIGHGKREGATLLTGGRPCGPEGKGYYIEPTVFTNVKEDMIIAKEEIFGPV 388
K QYE++L I GK+EGA L GG P G KGY+I+PTVF++V +DM IAKEEIFGPV
Sbjct: 347 KEQYEKILDLIESGKKEGAKLECGGGPWG--NKGYFIQPTVFSDVTDDMRIAKEEIFGPV 404
Query: 387 MCLMKFKPVEEAIARANDTRYGLGAGVVTRDLDXXXXXXXXXXXXXXXXNCYFAMGSDCP 208
+MKFK +++ I RAN+T YGL AG+ T D+D NCY + + CP
Sbjct: 405 QQIMKFKSLDDVIKRANNTFYGLSAGIFTNDIDKAITVSSALQSGTVWVNCYSVVSAQCP 464
Query: 207 FGGRKMSGFGKDEGMHALDKYLAVKSV 127
FGG KMSG G++ G + +Y VK+V
Sbjct: 465 FGGFKMSGNGRELGEYGFHEYTEVKTV 491
>sptr|Q43274|Q43274 T CYTOPLASM MALE sterility RESTORER factor 2 (EC
1.2.1.3) (Aldehyde dehydrogenase (NAD+)).
Length = 549
Score = 141 bits (356), Expect = 6e-33
Identities = 70/155 (45%), Positives = 100/155 (64%)
Frame = -1
Query: 561 QYERVLSYIGHGKREGATLLTGGRPCGPEGKGYYIEPTVFTNVKEDMIIAKEEIFGPVMC 382
Q+ ++L YI +G GATL+TGG G KG+YI+PT+F++V++ M IA+EEIFGPV
Sbjct: 397 QFNKILRYIRYGVDGGATLVTGGDRLGD--KGFYIQPTIFSDVQDGMKIAQEEIFGPVQS 454
Query: 381 LMKFKPVEEAIARANDTRYGLGAGVVTRDLDXXXXXXXXXXXXXXXXNCYFAMGSDCPFG 202
++KFK + E I RAN ++YGL AGV T LD NC+ + PFG
Sbjct: 455 ILKFKDLNEVIKRANASQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFG 514
Query: 201 GRKMSGFGKDEGMHALDKYLAVKSVVTPLRASPWI 97
G KMSG G+++G+ +L YL VK+VVTP++ + W+
Sbjct: 515 GYKMSGIGREKGVDSLKNYLQVKAVVTPIKNAAWL 549
>sptr|Q8QGQ2|Q8QGQ2 Aldehyde dehydrogenase 2.
Length = 516
Score = 141 bits (356), Expect = 6e-33
Identities = 72/145 (49%), Positives = 94/145 (64%)
Frame = -1
Query: 561 QYERVLSYIGHGKREGATLLTGGRPCGPEGKGYYIEPTVFTNVKEDMIIAKEEIFGPVMC 382
Q+++VL YI GKREGA L+ GG P +GY+I+PTVF +VK+DM IA+EEIFGPVM
Sbjct: 365 QFKKVLGYISSGKREGAKLMCGGAPAAE--RGYFIQPTVFGDVKDDMKIAREEIFGPVMQ 422
Query: 381 LMKFKPVEEAIARANDTRYGLGAGVVTRDLDXXXXXXXXXXXXXXXXNCYFAMGSDCPFG 202
++KFK +EE I RAND++YGL A V T+++D NCY G PFG
Sbjct: 423 ILKFKSLEEVIERANDSKYGLAAAVFTQNIDKANYISHGLRAGTVWINCYNVFGVQAPFG 482
Query: 201 GRKMSGFGKDEGMHALDKYLAVKSV 127
G K SG G++ G + LD Y VK+V
Sbjct: 483 GYKASGIGRELGEYGLDIYTEVKTV 507
>sw|Q8MI17|DHA1_RABIT Aldehyde dehydrogenase 1A1 (EC 1.2.1.3)
(Aldehyde dehydrogenase, cytosolic) (ALDH class 1).
Length = 496
Score = 141 bits (355), Expect = 7e-33
Identities = 72/151 (47%), Positives = 95/151 (62%)
Frame = -1
Query: 567 KAQYERVLSYIGHGKREGATLLTGGRPCGPEGKGYYIEPTVFTNVKEDMIIAKEEIFGPV 388
K QY ++L I GK+EGA L GG P G KGY+I+PTVF+NV ++M IAKEEIFGPV
Sbjct: 343 KEQYNKILDLIESGKKEGAKLECGGGPWG--NKGYFIQPTVFSNVTDEMRIAKEEIFGPV 400
Query: 387 MCLMKFKPVEEAIARANDTRYGLGAGVVTRDLDXXXXXXXXXXXXXXXXNCYFAMGSDCP 208
+MKFK +++ I RAN+T YGL AG+ T+DLD NCY + + P
Sbjct: 401 QQIMKFKSLDDVIKRANNTTYGLSAGIFTKDLDKAVTVSSALQAGTVWVNCYSVVSAQVP 460
Query: 207 FGGRKMSGFGKDEGMHALDKYLAVKSVVTPL 115
FGG KMSG G++ G + L +Y VK+V +
Sbjct: 461 FGGFKMSGNGRELGEYGLQQYTEVKTVTVKI 491
Database: /db/uniprot/tmp/swall
Posted date: Mar 5, 2004 7:45 PM
Number of letters in database: 442,889,342
Number of sequences in database: 1,395,590
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 388,050,683
Number of Sequences: 1395590
Number of extensions: 7494676
Number of successful extensions: 27241
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 24784
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25965
length of database: 442,889,342
effective HSP length: 117
effective length of database: 279,605,312
effective search space used: 23207240896
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

