BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= 2542042.2.1
(691 letters)
Database: /db/trembl-ebi/tmp/swall
1,272,877 sequences; 406,886,216 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
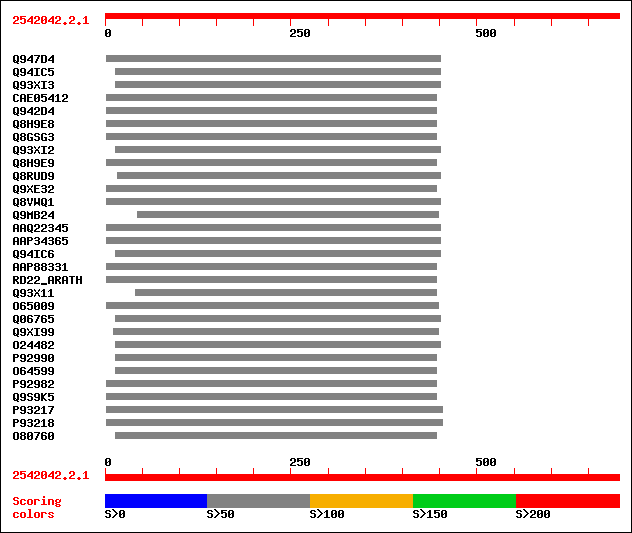
sptr|Q947D4|Q947D4 Dehydration-responsive protein RD22 (Fragment). 92 6e-18
sptr|Q94IC5|Q94IC5 BURP domain-containing protein (Fragment). 89 4e-17
sptr|Q93XI3|Q93XI3 BURP domain-containing protein. 89 7e-17
sptrnew|CAE05412|CAE05412 OSJNBa0036B17.16 protein. 88 9e-17
sptr|Q942D4|Q942D4 Putative dehydration-responsive protein RD22. 87 3e-16
sptr|Q8H9E8|Q8H9E8 Resistant specific protein-3. 87 3e-16
sptr|Q8GSG3|Q8GSG3 Resistant specific protein-1(4) (Resistant sp... 87 3e-16
sptr|Q93XI2|Q93XI2 BURP domain-containing protein (Fragment). 86 3e-16
sptr|Q8H9E9|Q8H9E9 Resistant specific protein-2. 84 1e-15
sptr|Q8RUD9|Q8RUD9 Seed coat BURP domain protein 1. 83 3e-15
sptr|Q9XE32|Q9XE32 EST AU029260(E30024) corresponds to a region ... 82 6e-15
sptr|Q8VWQ1|Q8VWQ1 Dehydration-induced protein RD22-like protein. 82 6e-15
sptr|Q9MB24|Q9MB24 S1-3 protein (Fragment). 82 8e-15
sptrnew|AAQ22345|AAQ22345 BURP domain-containing protein. 81 1e-14
sptrnew|AAP34365|AAP34365 Putative dehydration-induced protein (... 80 2e-14
sptr|Q94IC6|Q94IC6 BURP domain-containing protein (Fragment). 79 4e-14
sptrnew|AAP88331|AAP88331 At5g25610/T14C9_150. 77 2e-13
sw|Q08298|RD22_ARATH Dehydration-responsive protein RD22 precursor. 77 2e-13
sptr|Q93X11|Q93X11 A2-134 protein. 67 2e-10
sptr|O65009|O65009 BURP domain containing protein. 65 6e-10
sptr|Q06765|Q06765 ADR6 protein. 64 1e-09
sptr|Q9XI99|Q9XI99 Putative BURP domain containing protein (At1g... 60 3e-08
sptr|O24482|O24482 Sali3-2. 60 3e-08
sptr|P92990|P92990 Polygalacturonase isoenzyme 1 beta subunit ho... 59 4e-08
sptr|O64599|O64599 F17O7.9 protein. 59 4e-08
sptr|P92982|P92982 Aromatic rich glycoprotein JP630. 58 1e-07
sptr|Q9S9K5|Q9S9K5 F5O8.31 protein. 58 1e-07
sptr|P93217|P93217 AROGP2. 57 2e-07
sptr|P93218|P93218 AROGP3. 57 2e-07
sptr|O80760|O80760 T13D8.26 protein. 57 2e-07
>sptr|Q947D4|Q947D4 Dehydration-responsive protein RD22 (Fragment).
Length = 349
Score = 92.0 bits (227), Expect = 6e-18
Identities = 53/155 (34%), Positives = 83/155 (53%), Gaps = 5/155 (3%)
Frame = +2
Query: 2 LRTCEAES--PEAHACATSEQAAADFAASSLGVRASKLVALVTTVHGGKDATRYVVAPNG 175
+ CE+ E CATS ++ DF+ S LG + A+ T G +Y + P G
Sbjct: 200 IEECESSGIRGEEKYCATSLESMVDFSTSKLG---GNIQAISTEAEKGATLQKYTITP-G 255
Query: 176 ITRIGKAAGAAAVVPCHPMSYPYMVHYCHQPADVEALRIELTGLGDGGVTTAIAMCHANT 355
+ ++ AAG + V CH +YPY V YCH A + L G DG A+A+CH +T
Sbjct: 256 VKKL--AAGKSVV--CHKQTYPYAVFYCHATKTTRAYVVPLKG-ADGLEAKAVAVCHTDT 310
Query: 356 MNWDDRY--FQMLNVTRGE-EICHFMPRNYVLWLP 451
W+ ++ FQ+L V G +CHF+P+++++W+P
Sbjct: 311 SEWNPKHLAFQVLKVKPGTVPVCHFLPKDHIVWVP 345
>sptr|Q94IC5|Q94IC5 BURP domain-containing protein (Fragment).
Length = 346
Score = 89.4 bits (220), Expect = 4e-17
Identities = 56/153 (36%), Positives = 81/153 (52%), Gaps = 7/153 (4%)
Frame = +2
Query: 14 EAESP----EAHACATSEQAAADFAASSLGVRASKLVALVTTVHGGKDATRYVVAPNGIT 181
E E+P E CATS ++ DF+ S LG + A+ T V +Y +A
Sbjct: 203 ECENPGIEGEEKYCATSLESMVDFSTSKLG---KDIQAISTEVEKQTGRQQYTIAG---- 255
Query: 182 RIGKAAGAAAVVPCHPMSYPYMVHYCHQPADVEALRIELTGLGDGGVTTAIAMCHANTMN 361
+ K AG +VV CH +YPY V YCH A ++L G D A+A+CH NT
Sbjct: 256 -VKKTAGDKSVV-CHKQNYPYAVFYCHSTQSTRAYLVQLKG-ADRTRLQAVAICHTNTSA 312
Query: 362 WDDRY--FQMLNVTRGE-EICHFMPRNYVLWLP 451
W+ ++ FQ+L V G ICHF+P+++V+W+P
Sbjct: 313 WNPKHLAFQVLAVKPGTVPICHFLPQSHVVWVP 345
>sptr|Q93XI3|Q93XI3 BURP domain-containing protein.
Length = 336
Score = 88.6 bits (218), Expect = 7e-17
Identities = 56/153 (36%), Positives = 81/153 (52%), Gaps = 7/153 (4%)
Frame = +2
Query: 14 EAESP----EAHACATSEQAAADFAASSLGVRASKLVALVTTVHGGKDATRYVVAPNGIT 181
E E+P E CATS ++ DF+ S LG + A+ T V +Y +A
Sbjct: 193 ECENPGIEGEEKYCATSLESMVDFSTSKLG---KDIQAISTEVEKQTGRQKYTIAG---- 245
Query: 182 RIGKAAGAAAVVPCHPMSYPYMVHYCHQPADVEALRIELTGLGDGGVTTAIAMCHANTMN 361
+ K AG VV CH +YPY V YCH A ++L G+ D A+A+CH NT
Sbjct: 246 -VKKIAGDKCVV-CHKQNYPYAVFYCHSIQSTRAYLVQLKGV-DRTRLQAVAICHTNTSA 302
Query: 362 WDDRY--FQMLNVTRGE-EICHFMPRNYVLWLP 451
W+ ++ FQ+L V G ICHF+P+++V+W+P
Sbjct: 303 WNPKHPAFQVLAVKPGTVPICHFLPQSHVVWVP 335
>sptrnew|CAE05412|CAE05412 OSJNBa0036B17.16 protein.
Length = 398
Score = 88.2 bits (217), Expect = 9e-17
Identities = 56/154 (36%), Positives = 75/154 (48%), Gaps = 6/154 (3%)
Frame = +2
Query: 2 LRTCE--AESPEAHACATSEQAAADFAASSLGVRASKLVA-LVTTVHGGKDATRYVVAPN 172
L CE A E ACATS ++ DF ASSLG R K + + G A Y V
Sbjct: 247 LLECELPANKGEKKACATSLESMVDFVASSLGTRDIKAASTFLVGKDGDTPAQEYTV--- 303
Query: 173 GITRIGKAAGAAAVVPCHPMSYPYMVHYCHQPADVEALRIELTGLGDGGVTTAIAMCHAN 352
T + A ++ CHP SYPY V CH A + L G DG A+A+CH +
Sbjct: 304 --TGARRMAETGQLIACHPESYPYAVFMCHLTEATRAYKASLVG-KDGAAVEAVAVCHTD 360
Query: 353 TMNWDDRY--FQMLNVTRGE-EICHFMPRNYVLW 445
T W+ ++ FQ+L V G +CHF+ + V+W
Sbjct: 361 TAEWNPKHAAFQVLGVKPGTVPVCHFVQPDVVVW 394
>sptr|Q942D4|Q942D4 Putative dehydration-responsive protein RD22.
Length = 429
Score = 86.7 bits (213), Expect = 3e-16
Identities = 53/153 (34%), Positives = 76/153 (49%), Gaps = 5/153 (3%)
Frame = +2
Query: 2 LRTCEAESP--EAHACATSEQAAADFAASSLGVRASKLVALVTTVHGGKDATRYVVAPNG 175
LR CEA E ACATS ++ DFA SSLG + V +TV G + +
Sbjct: 277 LRDCEAPPAQGERKACATSLESMVDFATSSLG---TSHVRAASTVVGKEGSPEQEYTVTA 333
Query: 176 ITRIGKAAGAAAVVPCHPMSYPYMVHYCHQPADVEALRIELTGLGDGGVTTAIAMCHANT 355
+ R +V CH Y Y V CH A + + G DG A+A+CHA+T
Sbjct: 334 VKRAAAGGDQDQLVACHAEPYAYAVFACHLTRATRAYAVSMAG-RDGTGVEAVAVCHADT 392
Query: 356 MNWDDRY--FQMLNVTRGE-EICHFMPRNYVLW 445
W+ ++ FQ+L V G +CHF+P+++V+W
Sbjct: 393 AGWNPKHVAFQVLKVKPGTVPVCHFLPQDHVVW 425
>sptr|Q8H9E8|Q8H9E8 Resistant specific protein-3.
Length = 275
Score = 86.7 bits (213), Expect = 3e-16
Identities = 55/153 (35%), Positives = 77/153 (50%), Gaps = 5/153 (3%)
Frame = +2
Query: 2 LRTCE--AESPEAHACATSEQAAADFAASSLGVRASKLVALVTTVHGGKDATRYVVAPNG 175
L CE A E CATS ++ DF S LG A T + G +++V +G
Sbjct: 129 LNHCEKPALKGEEKQCATSVESMVDFVTSKLGNNAR---VTSTELEIGSKFQKFIVK-DG 184
Query: 176 ITRIGKAAGAAAVVPCHPMSYPYMVHYCHQPADVEALRIELTGLGDGGVTTAIAMCHANT 355
+ K ++ CHPMSYPY+V YCH+ A+ A + L G DG A+A+CH +T
Sbjct: 185 V----KILAEEKIIACHPMSYPYVVFYCHKMANSTAHFLPLEG-EDGTRVKAVAICHKDT 239
Query: 356 MNWDDRY--FQMLNVTRG-EEICHFMPRNYVLW 445
WD + FQ+L V G CHF P +++W
Sbjct: 240 SQWDPHHVAFQVLKVKPGTSSACHFFPEGHLVW 272
>sptr|Q8GSG3|Q8GSG3 Resistant specific protein-1(4) (Resistant
specific protein-1(8)).
Length = 402
Score = 86.7 bits (213), Expect = 3e-16
Identities = 55/153 (35%), Positives = 77/153 (50%), Gaps = 5/153 (3%)
Frame = +2
Query: 2 LRTCE--AESPEAHACATSEQAAADFAASSLGVRASKLVALVTTVHGGKDATRYVVAPNG 175
L CE A E CATS ++ DF S LG A T + G +++V +G
Sbjct: 256 LNHCEKPALKGEEKQCATSVESMVDFVTSKLGNNAR---VTSTELEIGSKFQKFIVK-DG 311
Query: 176 ITRIGKAAGAAAVVPCHPMSYPYMVHYCHQPADVEALRIELTGLGDGGVTTAIAMCHANT 355
+ K ++ CHPMSYPY+V YCH+ A+ A + L G DG A+A+CH +T
Sbjct: 312 V----KILAEEKIIACHPMSYPYVVFYCHKMANSTAHFLPLEG-EDGTRVKAVAICHKDT 366
Query: 356 MNWDDRY--FQMLNVTRG-EEICHFMPRNYVLW 445
WD + FQ+L V G CHF P +++W
Sbjct: 367 SQWDPHHVAFQVLKVKPGTSSACHFFPEGHLVW 399
>sptr|Q93XI2|Q93XI2 BURP domain-containing protein (Fragment).
Length = 307
Score = 86.3 bits (212), Expect = 3e-16
Identities = 55/153 (35%), Positives = 80/153 (52%), Gaps = 7/153 (4%)
Frame = +2
Query: 14 EAESP----EAHACATSEQAAADFAASSLGVRASKLVALVTTVHGGKDATRYVVAPNGIT 181
E E+P E CATS ++ DF+ S LG + A+ T V +Y +A
Sbjct: 164 ECENPGIEGEEKYCATSLESMVDFSTSKLG---KDVQAISTEVEKQTGRQKYTIAG---- 216
Query: 182 RIGKAAGAAAVVPCHPMSYPYMVHYCHQPADVEALRIELTGLGDGGVTTAIAMCHANTMN 361
+ K AG VV CH +YPY V YCH A ++L G+ D A+A+CH NT
Sbjct: 217 -VKKIAGDKCVV-CHKQNYPYAVFYCHSIQSTRAYLVQLKGV-DRTRLQAVAICHTNTSA 273
Query: 362 WDDRY--FQMLNVTRGE-EICHFMPRNYVLWLP 451
W+ ++ FQ+L V G CHF+P+++V+W+P
Sbjct: 274 WNPKHPAFQVLAVKPGTVPTCHFLPQSHVVWVP 306
>sptr|Q8H9E9|Q8H9E9 Resistant specific protein-2.
Length = 440
Score = 84.3 bits (207), Expect = 1e-15
Identities = 53/153 (34%), Positives = 76/153 (49%), Gaps = 5/153 (3%)
Frame = +2
Query: 2 LRTCE--AESPEAHACATSEQAAADFAASSLGVRASKLVALVTTVHGGKDATRYVVAPNG 175
L CE A E CATS ++ DF S LG A + +T + + + +G
Sbjct: 294 LNNCEEPALKGEEKHCATSVESMVDFVTSKLGNNAR----VTSTELEIESKFQKFIVKDG 349
Query: 176 ITRIGKAAGAAAVVPCHPMSYPYMVHYCHQPADVEALRIELTGLGDGGVTTAIAMCHANT 355
+ K ++ CHPMSYPY+V YCH+ ++ A + L G DG AI +CH +T
Sbjct: 350 V----KILAEEEIIACHPMSYPYVVFYCHKMSNSTAHVVPLEG-EDGTRVKAIVICHKDT 404
Query: 356 MNWDDRY--FQMLNVTRG-EEICHFMPRNYVLW 445
WD + FQ+L V G +CHF P ++LW
Sbjct: 405 SQWDPDHVAFQVLKVKPGTSPVCHFFPNGHLLW 437
>sptr|Q8RUD9|Q8RUD9 Seed coat BURP domain protein 1.
Length = 305
Score = 83.2 bits (204), Expect = 3e-15
Identities = 50/148 (33%), Positives = 71/148 (47%), Gaps = 3/148 (2%)
Frame = +2
Query: 17 AESPEAHACATSEQAAADFAASSLGVRASKLVALVTTVHGGKDATRYVVAPNGITRIGKA 196
A E CATS ++ DF S+LG A T ++ ++VV NG+ ++G
Sbjct: 165 ATEGERKHCATSLESMVDFVVSALGKNVG---AFSTEKERETESGKFVVVKNGVRKLGDD 221
Query: 197 AGAAAVVPCHPMSYPYMVHYCHQPADVEALRIELTGLGDGGVTTAIAMCHANTMNWDDRY 376
V+ CHPMSYPY+V CH + L G DG A+ CH +T WD +
Sbjct: 222 K----VIACHPMSYPYVVFGCHLVPRSSGYLVRLKG-EDGVRVKAVVACHRDTSKWDHNH 276
Query: 377 --FQMLNVTRGE-EICHFMPRNYVLWLP 451
F++LN+ G +CH +LWLP
Sbjct: 277 GAFKVLNLKPGNGTVCHVFTEGNLLWLP 304
>sptr|Q9XE32|Q9XE32 EST AU029260(E30024) corresponds to a region of
the predicted gene.
Length = 413
Score = 82.0 bits (201), Expect = 6e-15
Identities = 54/157 (34%), Positives = 77/157 (49%), Gaps = 9/157 (5%)
Frame = +2
Query: 2 LRTCE--AESPEAHACATSEQAAADFAASSLGVRASKLVALVTTV-HGGKDATRYVVAPN 172
LRTCE + E+ CATS +A + A ++LG R + AL +T+ GG Y V
Sbjct: 256 LRTCEWPTLAGESKFCATSLEALVEGAMAALGTR--DIAALASTLPRGGAPLQAYAV--- 310
Query: 173 GITRIGKAAGAAAVVPCHPMSYPYMVHYCHQPADVEALRIEL--TGLGDGGVTTAIA-MC 343
R A V CH +YPY V+ CH A +E+ G GDGG +A +C
Sbjct: 311 ---RAVLPVEGAGFVACHDQAYPYTVYRCHTTGPARAYMVEMEGDGGGDGGEAVTVATVC 367
Query: 344 HANTMNWDDRY--FQMLNV-TRGEEICHFMPRNYVLW 445
H NT W+ + F++L G +CH MP +++W
Sbjct: 368 HTNTSRWNPEHVSFKLLGTKPGGSPVCHLMPYGHIVW 404
>sptr|Q8VWQ1|Q8VWQ1 Dehydration-induced protein RD22-like protein.
Length = 335
Score = 82.0 bits (201), Expect = 6e-15
Identities = 51/155 (32%), Positives = 79/155 (50%), Gaps = 5/155 (3%)
Frame = +2
Query: 2 LRTCEAESPEAHA--CATSEQAAADFAASSLGVRASKLVALVTTVHGGKDATRYVVAPNG 175
++ CE + E CATS ++ D++ S LG A+ T V +Y +A
Sbjct: 189 IKECEQPAIEGEEKYCATSLESMIDYSISKLGKVDQ---AVSTEVEKQTPMQKYTIAAG- 244
Query: 176 ITRIGKAAGAAAVVPCHPMSYPYMVHYCHQPADVEALRIELTGLGDGGVTTAIAMCHANT 355
+ K AVV CH +Y Y V YCH+ A + L G DG A+A+CH +T
Sbjct: 245 ---VQKMTDDKAVV-CHKQNYAYAVFYCHKSETTRAYMVPLEG-ADGTKAKAVAVCHTDT 299
Query: 356 MNWDDRY--FQMLNVTRGE-EICHFMPRNYVLWLP 451
W+ ++ FQ+L V G +CHF+PR++++W+P
Sbjct: 300 SAWNPKHLAFQVLKVEPGTIPVCHFLPRDHIVWVP 334
>sptr|Q9MB24|Q9MB24 S1-3 protein (Fragment).
Length = 132
Score = 81.6 bits (200), Expect = 8e-15
Identities = 48/138 (34%), Positives = 73/138 (52%), Gaps = 3/138 (2%)
Frame = +2
Query: 44 ATSEQAAADFAASSLGVRASKLVALVTTVHGGKDATRYVVAPNGITRIGKAAGAAAVVPC 223
ATS ++ DF+ S LG + L T V +Y +AP + K +G AVV C
Sbjct: 2 ATSLESMVDFSTSKLG---KNVAVLSTEVDQETGLQQYTIAPG----VKKVSGDNAVV-C 53
Query: 224 HPMSYPYMVHYCHQPADVEALRIELTGLGDGGVTTAIAMCHANTMNWDDRY--FQMLNVT 397
H SYPY V YCH+ + L G +G A+A+CH +T W+ ++ F++L V
Sbjct: 54 HKQSYPYAVFYCHKTETTRTYPVPLEG-ANGIRVKAVAVCHTDTSQWNPKHLAFEVLKVK 112
Query: 398 RGE-EICHFMPRNYVLWL 448
G +CHF+P ++V+W+
Sbjct: 113 PGTVPVCHFLPEDHVVWV 130
>sptrnew|AAQ22345|AAQ22345 BURP domain-containing protein.
Length = 335
Score = 80.9 bits (198), Expect = 1e-14
Identities = 51/155 (32%), Positives = 78/155 (50%), Gaps = 5/155 (3%)
Frame = +2
Query: 2 LRTCEAESPEAHA--CATSEQAAADFAASSLGVRASKLVALVTTVHGGKDATRYVVAPNG 175
++ CE + E CATS ++ D++ S LG A+ T V +Y +A
Sbjct: 189 IKECEQPAIEGEEKYCATSLESMIDYSISKLGKVDQ---AVSTEVEKQTPMQKYTIAAG- 244
Query: 176 ITRIGKAAGAAAVVPCHPMSYPYMVHYCHQPADVEALRIELTGLGDGGVTTAIAMCHANT 355
+ K AVV CH +Y Y V YCH+ A + L G G G A+A+CH +T
Sbjct: 245 ---VQKMTDDKAVV-CHKQNYAYAVFYCHKSETTRAYMVPLEGAG-GTKAQALAVCHTDT 299
Query: 356 MNWDDRY--FQMLNVTRGE-EICHFMPRNYVLWLP 451
W+ ++ FQ L V G +CHF+PR++++W+P
Sbjct: 300 SAWNPKHLAFQFLKVEPGTIPVCHFLPRDHIVWVP 334
>sptrnew|AAP34365|AAP34365 Putative dehydration-induced protein
(Fragment).
Length = 156
Score = 80.5 bits (197), Expect = 2e-14
Identities = 50/155 (32%), Positives = 78/155 (50%), Gaps = 5/155 (3%)
Frame = +2
Query: 2 LRTCEAESPEAHA--CATSEQAAADFAASSLGVRASKLVALVTTVHGGKDATRYVVAPNG 175
++ CE + E CATS ++ D++ S LG A+ T V +Y +A
Sbjct: 10 IKECEQPAIEGEEKYCATSLESMIDYSISKLGKVDQ---AVSTEVEKQTPMQKYTIAAG- 65
Query: 176 ITRIGKAAGAAAVVPCHPMSYPYMVHYCHQPADVEALRIELTGLGDGGVTTAIAMCHANT 355
+ K AVV CH +Y Y V YCH+ A + L G DG A+A+CH +T
Sbjct: 66 ---VQKMTDDKAVV-CHKQNYAYAVFYCHKSETTRAYMVPLEG-ADGTKAKAVAVCHTDT 120
Query: 356 MNWDDRY--FQMLNVTRGE-EICHFMPRNYVLWLP 451
W+ ++ FQ+L G +CHF+PR++++W+P
Sbjct: 121 SAWNPKHLAFQVLKAEPGTVPVCHFLPRDHIVWVP 155
>sptr|Q94IC6|Q94IC6 BURP domain-containing protein (Fragment).
Length = 292
Score = 79.3 bits (194), Expect = 4e-14
Identities = 53/153 (34%), Positives = 78/153 (50%), Gaps = 7/153 (4%)
Frame = +2
Query: 14 EAESP----EAHACATSEQAAADFAASSLGVRASKLVALVTTVHGGKDATRYVVAPNGIT 181
E E+P E C TS ++ DF++S LG + A+ T V +Y +A
Sbjct: 150 ECENPRIEGEEKYCVTSLESIVDFSSSKLG---KDIQAISTEVEKQTGRQKYTIAV---- 202
Query: 182 RIGKAAGAAAVVPCHPMSYPYMVHYCHQPADVEALRIELTGLGDGGVTTAIAMCHANTMN 361
+ K AG +VV CH +YPY V YCH A ++L G D A+A CH NT
Sbjct: 203 -VKKMAGDKSVV-CHKQNYPYAVFYCH-TTQSRAYLVQLKG-ADRSELKAVATCHTNTSA 258
Query: 362 WDDRY--FQMLNVTRGE-EICHFMPRNYVLWLP 451
W+ ++ FQ+L V G +CHF+ +++V+W P
Sbjct: 259 WNPKHMAFQVLAVKPGTVPVCHFLTQSHVVWAP 291
>sptrnew|AAP88331|AAP88331 At5g25610/T14C9_150.
Length = 392
Score = 77.0 bits (188), Expect = 2e-13
Identities = 51/155 (32%), Positives = 76/155 (49%), Gaps = 7/155 (4%)
Frame = +2
Query: 2 LRTCEAE--SPEAHACATSEQAAADFAASSLGVRASKLVALVTTVHGGKDAT--RYVVAP 169
+ CEA S E CATS ++ DF+ S LG V V+T K+A +Y +A
Sbjct: 243 IEECEARKVSGEEKYCATSLESMVDFSVSKLG---KYHVRAVSTEVAKKNAPMQKYKIAA 299
Query: 170 NGITRIGKAAGAAAVVPCHPMSYPYMVHYCHQPADVEALRIELTGLGDGGVTTAIAMCHA 349
G+ ++ V CH YP+ V YCH+ + L G +G A+A+CH
Sbjct: 300 AGVKKLSDDKS----VVCHKQKYPFAVFYCHKAMMTTVYAVPLEG-ENGMRAKAVAVCHK 354
Query: 350 NTMNWDDRY--FQMLNVTRGE-EICHFMPRNYVLW 445
NT W+ + F++L V G +CHF+P +V+W
Sbjct: 355 NTSAWNPNHLAFKVLKVKPGTVPVCHFLPETHVVW 389
>sw|Q08298|RD22_ARATH Dehydration-responsive protein RD22 precursor.
Length = 392
Score = 77.0 bits (188), Expect = 2e-13
Identities = 51/155 (32%), Positives = 76/155 (49%), Gaps = 7/155 (4%)
Frame = +2
Query: 2 LRTCEAE--SPEAHACATSEQAAADFAASSLGVRASKLVALVTTVHGGKDAT--RYVVAP 169
+ CEA S E CATS ++ DF+ S LG V V+T K+A +Y +A
Sbjct: 243 IEECEARKVSGEEKYCATSLESMVDFSVSKLG---KYHVRAVSTEVAKKNAPMQKYKIAA 299
Query: 170 NGITRIGKAAGAAAVVPCHPMSYPYMVHYCHQPADVEALRIELTGLGDGGVTTAIAMCHA 349
G+ ++ V CH YP+ V YCH+ + L G +G A+A+CH
Sbjct: 300 AGVKKLSDDKS----VVCHKQKYPFAVFYCHKAMMTTVYAVPLEG-ENGMRAKAVAVCHK 354
Query: 350 NTMNWDDRY--FQMLNVTRGE-EICHFMPRNYVLW 445
NT W+ + F++L V G +CHF+P +V+W
Sbjct: 355 NTSAWNPNHLAFKVLKVKPGTVPVCHFLPETHVVW 389
>sptr|Q93X11|Q93X11 A2-134 protein.
Length = 305
Score = 67.0 bits (162), Expect = 2e-10
Identities = 44/139 (31%), Positives = 66/139 (47%), Gaps = 4/139 (2%)
Frame = +2
Query: 41 CATSEQAAADFAASSLGVRASKLVALVTTVHGGKDATRYVVAPNGITRIGKAAGAAAVVP 220
C TS AA+ LG +++A + GG Y V R K + V
Sbjct: 167 CVTSPDDMVGRAAAVLGTSNMQVLA-PSIPTGGMSLQPYTV------RAVKPVDGSDFVG 219
Query: 221 CHPMSYPYMVHYCHQPADVEALRIEL-TGLGDGGVTTAIAMCHANTMNWDDRY--FQML- 388
CHP YPY V+ CH +E+ + G+GG +A+CH NT +WD + F++L
Sbjct: 220 CHPELYPYSVYRCHTSVQTGTYVMEMQSSYGNGGALKLVAVCHRNTTSWDPEHVSFKVLA 279
Query: 389 NVTRGEEICHFMPRNYVLW 445
+ G ICHF+P +V++
Sbjct: 280 SKPGGLPICHFVPYGHVIF 298
>sptr|O65009|O65009 BURP domain containing protein.
Length = 282
Score = 65.5 bits (158), Expect = 6e-10
Identities = 40/157 (25%), Positives = 72/157 (45%), Gaps = 8/157 (5%)
Frame = +2
Query: 2 LRTCEAESPEAHA--CATSEQAAADFAASSLGVRASKLVALVTTVHGGKDATRYVVAPNG 175
L+ C+ ++ E C TS ++ D A ++ A V + ++ Y +
Sbjct: 126 LKRCDFKAIEGEYKFCGTSLESMLDLAKKTIASNADLKVMTTKVMVPDQNRISYALHNYT 185
Query: 176 ITRIGKAAGAAAVVPCHPMSYPYMVHYCH-QPADVEALRIELTGLGDGGVTTAI--AMCH 346
+ K V+ CH M YPY+V+YCH + + + L + D G+ + A+CH
Sbjct: 186 FAEVPKELDGIKVLGCHRMPYPYVVYYCHGHKSGTKVFEVNL--MSDDGIQLVVGPAVCH 243
Query: 347 ANTMNW--DDRYFQMLNV-TRGEEICHFMPRNYVLWL 448
+T W D F++L + R +CHF P + ++W+
Sbjct: 244 MDTSMWNADHVAFKVLKIEPRSAPVCHFFPLDNIVWV 280
>sptr|Q06765|Q06765 ADR6 protein.
Length = 272
Score = 64.3 bits (155), Expect = 1e-09
Identities = 46/148 (31%), Positives = 71/148 (47%), Gaps = 2/148 (1%)
Frame = +2
Query: 14 EAESPEAHACATSEQAAADFAASSLGVRASKLVALVTTVHGGKDATRYVVAPNGITRIGK 193
EA E CA S FA S LG K + ++++ K +Y V G+ +G
Sbjct: 124 EAIEGEEKFCAKSLGTVIGFAISKLG----KNIQVLSSSFVNKQ-DQYTV--EGVQNLGD 176
Query: 194 AAGAAAVVPCHPMSYPYMVHYCHQPADVEALRIELTGLGDGGVTTAIAMCHANTMNWDDR 373
A V CH +++ V YCH+ + A + L GDG T A+A+CH+NT + +
Sbjct: 177 KA-----VMCHRLNFRTAVFYCHEVRETTAFMVPLVA-GDGTKTQALAICHSNTSGMNHQ 230
Query: 374 YF-QMLNVTRG-EEICHFMPRNYVLWLP 451
Q++ V G +CHF+ +LW+P
Sbjct: 231 MLHQLMGVDPGTNPVCHFLGSKAILWVP 258
>sptr|Q9XI99|Q9XI99 Putative BURP domain containing protein
(At1g49320).
Length = 280
Score = 60.1 bits (144), Expect = 3e-08
Identities = 38/153 (24%), Positives = 68/153 (44%), Gaps = 7/153 (4%)
Frame = +2
Query: 11 CEAESPEAHA--CATSEQAAADFAASSLGVRASKLVALVTTVHGGKDATRYVVAPNGITR 184
C+A++ E C TS ++ D ++G V + +++ Y +
Sbjct: 127 CDAKAIEGEHKFCGTSLESLIDLVKKTMGYNVDLKVMTTKVMVPAQNSISYALHNYTFVE 186
Query: 185 IGKAAGAAAVVPCHPMSYPYMVHYCHQPADVEALRIELTGLGDGGVTTAI--AMCHANTM 358
K ++ CH M YPY V+YCH + E+ + D G + A+CH +T
Sbjct: 187 APKELVGIKMLGCHRMPYPYAVYYCHGHKGGSRV-FEVNLVTDDGRQRVVGPAVCHMDTS 245
Query: 359 NWDDRY--FQMLNV-TRGEEICHFMPRNYVLWL 448
WD + F++L + R +CHF P + ++W+
Sbjct: 246 TWDADHVAFKVLKMEPRSAPVCHFFPLDNIVWV 278
>sptr|O24482|O24482 Sali3-2.
Length = 276
Score = 59.7 bits (143), Expect = 3e-08
Identities = 44/148 (29%), Positives = 70/148 (47%), Gaps = 2/148 (1%)
Frame = +2
Query: 14 EAESPEAHACATSEQAAADFAASSLGVRASKLVALVTTVHGGKDATRYVVAPNGITRIGK 193
EA E CA S FA S LG K + ++++ K +Y V G+ +G
Sbjct: 128 EAFEGEEKFCAKSLGTVIGFAISKLG----KNIQVLSSSFVNKQE-QYTV--EGVQNLGD 180
Query: 194 AAGAAAVVPCHPMSYPYMVHYCHQPADVEALRIELTGLGDGGVTTAIAMCHANTMNWDDR 373
A V CH +++ V YCH+ + A + L GDG T A+A+CH++T +
Sbjct: 181 KA-----VMCHGLNFRTAVFYCHKVRETTAFVVPLVA-GDGTKTQALAVCHSDTSGMNHH 234
Query: 374 YF-QMLNVTRG-EEICHFMPRNYVLWLP 451
+++ V G +CHF+ +LW+P
Sbjct: 235 ILHELMGVDPGTNPVCHFLGSKAILWVP 262
>sptr|P92990|P92990 Polygalacturonase isoenzyme 1 beta subunit
homolog.
Length = 626
Score = 59.3 bits (142), Expect = 4e-08
Identities = 49/153 (32%), Positives = 67/153 (43%), Gaps = 9/153 (5%)
Frame = +2
Query: 14 EAESP----EAHACATSEQAAADFAASSLGVRASKLVALVTTVHGGKDATRYVVAP-NGI 178
E E P E C S + DFA S LG + V L TT + + V+ NGI
Sbjct: 478 ECERPPSVGETKRCVGSAEDMIDFATSVLG----RSVVLRTTENVAGSKEKVVIGKVNGI 533
Query: 179 TRIGKAAGAAAVVPCHPMSYPYMVHYCHQPADVEALRIELTGLGD-GGVTTAIAMCHANT 355
GK A V CH YPY+++YCH V +L L + IA+CH +T
Sbjct: 534 NG-GKLTKA---VSCHQSLYPYLLYYCHSVPKVRVYEADLLELNSKKKINHGIAICHMDT 589
Query: 356 MNWDDRY--FQMLNVTRGE-EICHFMPRNYVLW 445
+W + F L G E+CH++ N + W
Sbjct: 590 SSWGPSHGAFLALGSKPGRIEVCHWIFENDMNW 622
>sptr|O64599|O64599 F17O7.9 protein.
Length = 626
Score = 59.3 bits (142), Expect = 4e-08
Identities = 49/153 (32%), Positives = 67/153 (43%), Gaps = 9/153 (5%)
Frame = +2
Query: 14 EAESP----EAHACATSEQAAADFAASSLGVRASKLVALVTTVHGGKDATRYVVAP-NGI 178
E E P E C S + DFA S LG + V L TT + + V+ NGI
Sbjct: 478 ECERPPSVGETKRCVGSAEDMIDFATSVLG----RSVVLRTTENVAGSKEKVVIGKVNGI 533
Query: 179 TRIGKAAGAAAVVPCHPMSYPYMVHYCHQPADVEALRIELTGLGD-GGVTTAIAMCHANT 355
GK A V CH YPY+++YCH V +L L + IA+CH +T
Sbjct: 534 NG-GKLTKA---VSCHQSLYPYLLYYCHSVPKVRVYEADLLELNSKKKINHGIAICHMDT 589
Query: 356 MNWDDRY--FQMLNVTRGE-EICHFMPRNYVLW 445
+W + F L G E+CH++ N + W
Sbjct: 590 SSWGPSHGAFLALGSKPGRIEVCHWIFENDMNW 622
>sptr|P92982|P92982 Aromatic rich glycoprotein JP630.
Length = 622
Score = 58.2 bits (139), Expect = 1e-07
Identities = 46/156 (29%), Positives = 69/156 (44%), Gaps = 8/156 (5%)
Frame = +2
Query: 2 LRTCEAESP--EAHACATSEQAAADFAASSLGVRASKLVALVTTVHGGKDATRYVVAPNG 175
+R CE E C S + DFA S LG S ++ +V G K+ + NG
Sbjct: 472 VRECERPPTVSETKRCVGSAEDMIDFATSVLG--RSVVLRTTESVAGSKEKVM-IGKVNG 528
Query: 176 IT--RIGKAAGAAAVVPCHPMSYPYMVHYCHQPADVEALRIELTG-LGDGGVTTAIAMCH 346
I R+ K+ V CH YPY+++YCH V +L + IA+CH
Sbjct: 529 INGGRVTKS------VSCHQSLYPYLLYYCHSVPKVRVYESDLLDPKSKAKINHGIAICH 582
Query: 347 ANTMNWDDRY--FQMLNVTRGE-EICHFMPRNYVLW 445
+T W + F +L G+ E+CH++ N + W
Sbjct: 583 MDTSAWGANHGAFMLLGSRPGQIEVCHWIFENDMNW 618
>sptr|Q9S9K5|Q9S9K5 F5O8.31 protein.
Length = 622
Score = 58.2 bits (139), Expect = 1e-07
Identities = 46/156 (29%), Positives = 69/156 (44%), Gaps = 8/156 (5%)
Frame = +2
Query: 2 LRTCEAESP--EAHACATSEQAAADFAASSLGVRASKLVALVTTVHGGKDATRYVVAPNG 175
+R CE E C S + DFA S LG S ++ +V G K+ + NG
Sbjct: 472 VRECERPPTVSETKRCVGSAEDMIDFATSVLG--RSVVLRTTESVAGSKEKVM-IGKVNG 528
Query: 176 IT--RIGKAAGAAAVVPCHPMSYPYMVHYCHQPADVEALRIELTG-LGDGGVTTAIAMCH 346
I R+ K+ V CH YPY+++YCH V +L + IA+CH
Sbjct: 529 INGGRVTKS------VSCHQSLYPYLLYYCHSVPKVRVYESDLLDPKSKAKINHGIAICH 582
Query: 347 ANTMNWDDRY--FQMLNVTRGE-EICHFMPRNYVLW 445
+T W + F +L G+ E+CH++ N + W
Sbjct: 583 MDTSAWGANHGAFMLLGSRPGQIEVCHWIFENDMNW 618
>sptr|P93217|P93217 AROGP2.
Length = 629
Score = 57.4 bits (137), Expect = 2e-07
Identities = 45/157 (28%), Positives = 66/157 (42%), Gaps = 6/157 (3%)
Frame = +2
Query: 2 LRTCE-AESP-EAHACATSEQAAADFAASSLGVRASKLVALVTTVHGGKDATRYVVAPNG 175
L CE A SP E C S + DFA S LG +V T G + + G
Sbjct: 479 LSECERAPSPGETKQCVNSAEDMIDFATSVLG---RNVVVRTTENTNGSKGNIMIGSIKG 535
Query: 176 ITRIGKAAGAAAVVPCHPMSYPYMVHYCHQPADVEALRIE-LTGLGDGGVTTAIAMCHAN 352
I GK + V CH YP +++YCH V + L + +A+CH +
Sbjct: 536 ING-GKVTKS---VSCHQTLYPSLLYYCHSVPKVRVYEADILDPNSKAKINHGVAICHVD 591
Query: 353 TMNWDDRYFQMLNVTRGE---EICHFMPRNYVLWLPA 454
T +W R+ + + G E+CH++ N + W A
Sbjct: 592 TSSWGPRHGAFIALGSGPGKIEVCHWIFENDMTWATA 628
>sptr|P93218|P93218 AROGP3.
Length = 632
Score = 57.0 bits (136), Expect = 2e-07
Identities = 45/157 (28%), Positives = 66/157 (42%), Gaps = 6/157 (3%)
Frame = +2
Query: 2 LRTCE-AESP-EAHACATSEQAAADFAASSLGVRASKLVALVTTVHGGKDATRYVVAPNG 175
L CE A SP E C S + DFA S LG +V T G + + G
Sbjct: 482 LSECERAPSPGETKQCVNSAEDMIDFATSVLG---RNVVVRTTENTNGSKGNIMIGSIKG 538
Query: 176 ITRIGKAAGAAAVVPCHPMSYPYMVHYCHQPADVEALRIE-LTGLGDGGVTTAIAMCHAN 352
I GK + V CH YP +++YCH V + L + +A+CH +
Sbjct: 539 ING-GKVTKS---VSCHQTLYPSLLYYCHSVPKVRVYEADILDPNSKAKINHGVAICHVD 594
Query: 353 TMNWDDRYFQMLNVTRGE---EICHFMPRNYVLWLPA 454
T +W R+ + + G E+CH++ N + W A
Sbjct: 595 TSSWGPRHGAFVALGSGPGKIEVCHWIFENDMTWATA 631
>sptr|O80760|O80760 T13D8.26 protein.
Length = 624
Score = 57.0 bits (136), Expect = 2e-07
Identities = 45/155 (29%), Positives = 64/155 (41%), Gaps = 11/155 (7%)
Frame = +2
Query: 14 EAESPEAHA----CATSEQAAADFAASSLGVRASKLVALVTTVHGGKDATRYVVAPNGIT 181
E E P +H C S + DFA S LG +V V G K + NGI
Sbjct: 476 ECERPASHGETKRCVGSAEDMIDFATSVLG--RGVVVRTTENVVGSKKKV-VIGKVNGIN 532
Query: 182 RIGKAAGAAAVVPCHPMSYPYMVHYCHQPADVEALRIELTGLGD----GGVTTAIAMCHA 349
V CH YPY+++YCH V +R+ T L D + +A+CH
Sbjct: 533 ----GGDVTRAVSCHQSLYPYLLYYCH---SVPRVRVYETDLLDPKSLEKINHGVAICHI 585
Query: 350 NTMNWDDRYFQMLNVTRGE---EICHFMPRNYVLW 445
+T W + L + G E+CH++ N + W
Sbjct: 586 DTSAWSPSHGAFLALGSGPGQIEVCHWIFENDMTW 620
Database: /db/trembl-ebi/tmp/swall
Posted date: Sep 26, 2003 8:29 PM
Number of letters in database: 406,886,216
Number of sequences in database: 1,272,877
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 422,757,841
Number of Sequences: 1272877
Number of extensions: 7858967
Number of successful extensions: 27628
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 26587
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 27550
length of database: 406,886,216
effective HSP length: 119
effective length of database: 255,413,853
effective search space used: 28095523830
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

