BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= 2441114.2.2
(962 letters)
Database: /db/trembl-ebi/tmp/swall
1,272,877 sequences; 406,886,216 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
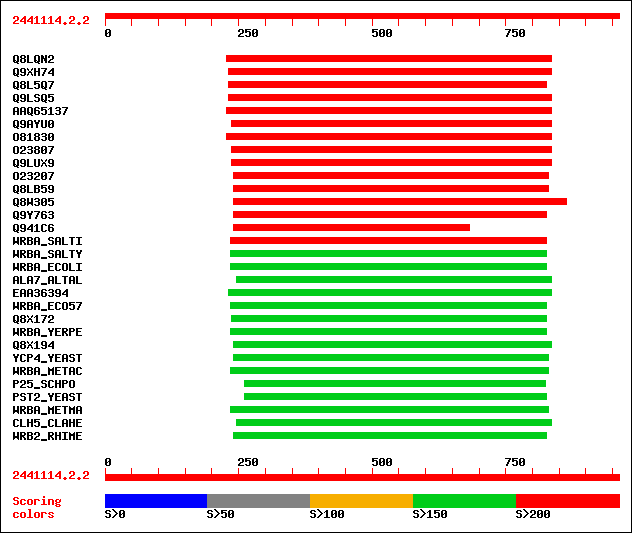
sptr|Q8LQN2|Q8LQN2 Putative 1,4-benzoquinone reductase. 385 e-106
sptr|Q9XH74|Q9XH74 Hypothetical 21.7 kDa protein. 344 1e-93
sptr|Q8L5Q7|Q8L5Q7 Putative quinone oxidoreductase (EC 1.6.5.5). 343 2e-93
sptr|Q9LSQ5|Q9LSQ5 1,4-benzoquinone reductase-like, Trp represso... 335 7e-91
sptrnew|AAQ65137|AAQ65137 At4g27270. 333 2e-90
sptr|Q9AYU0|Q9AYU0 Quinone-oxidoreductase QR2. 325 8e-88
sptr|O81830|O81830 Hypothetical 22.3 kDa protein. 310 1e-83
sptr|O23807|O23807 LEDI-3 protein. 295 5e-79
sptr|Q9LUX9|Q9LUX9 1,4-benzoquinone reductase-like (Putative lig... 273 3e-72
sptr|O23207|O23207 Minor allergen. 270 2e-71
sptr|Q8LB59|Q8LB59 Minor allergen. 270 2e-71
sptr|Q8W305|Q8W305 Putative reductase. 238 9e-62
sptr|Q9Y763|Q9Y763 1,4-benzoquinone reductase. 228 1e-58
sptr|Q941C6|Q941C6 C7A10_610/C7A10_610 (At4g36690/C7A10_610). 212 7e-54
sw|Q8Z7N9|WRBA_SALTI Flavoprotein wrbA (Trp repressor binding pr... 201 1e-50
sw|Q8ZQ40|WRBA_SALTY Flavoprotein wrbA (Trp repressor binding pr... 200 3e-50
sw|P30849|WRBA_ECOLI Flavoprotein wrbA (Trp repressor binding pr... 199 4e-50
sw|P42058|ALA7_ALTAL Minor allergen Alt a 7 (Alt a VII). 198 1e-49
sptrnew|EAA36394|EAA36394 Hypothetical protein. 196 4e-49
sw|Q8X4B4|WRBA_ECO57 Flavoprotein wrbA (Trp repressor binding pr... 196 5e-49
sptr|Q8X172|Q8X172 NADH:quinone oxidoreductase. 196 5e-49
sw|Q8ZF61|WRBA_YERPE Flavoprotein wrbA (Trp repressor binding pr... 194 2e-48
sptr|Q8X194|Q8X194 Y20 protein. 193 3e-48
sw|P25349|YCP4_YEAST Hypothetical 26.4 kDa protein in CDC10-CIT2... 192 8e-48
sw|P58796|WRBA_METAC Flavoprotein wrbA. 181 1e-44
sw|P30821|P25_SCHPO P25 protein (Brefeldin A resistance protein). 180 3e-44
sw|Q12335|PST2_YEAST Protoplast secreted protein 2 precursor. 179 7e-44
sw|Q8PUV4|WRBA_METMA Flavoprotein wrbA. 174 2e-42
sw|P42059|CLH5_CLAHE Minor allergen Cla h 5 (Cla h V). 165 1e-39
sw|Q930L2|WRB2_RHIME Flavoprotein wrbA2. 162 5e-39
>sptr|Q8LQN2|Q8LQN2 Putative 1,4-benzoquinone reductase.
Length = 203
Score = 385 bits (988), Expect = e-106
Identities = 185/203 (91%), Positives = 193/203 (95%)
Frame = -1
Query: 836 MAVKVYVVYYSMYGHVGKLAEEIQKGASSVEGVEAKTWQVPETLPEEVLGKMGAPPKPDV 657
MAVKVYVVYYSMYGHV KLAEEI+KGASS+EGVEAK WQVPETL EEVLGKMGAPPKPDV
Sbjct: 1 MAVKVYVVYYSMYGHVAKLAEEIKKGASSIEGVEAKIWQVPETLHEEVLGKMGAPPKPDV 60
Query: 656 PVITPQELAEADGILFGFPTRFGMMAAQMKAFFDATGGLWREQSLAGKPAGVFFCTGSQG 477
P ITPQEL EADGILFGFPTRFGMMAAQMKAFFDATGGLW EQSLAGKPAG+FF TG+QG
Sbjct: 61 PTITPQELTEADGILFGFPTRFGMMAAQMKAFFDATGGLWSEQSLAGKPAGIFFSTGTQG 120
Query: 476 GGQETTALTAITQLTHHGMVFVPVGYTFGAKLFGMDQVQGGSPYGAGTFAADGSRWPSEV 297
GGQETT LTAITQLTHHGMVFVPVGYTFGAK+F M +VQGGSPYGAGTFAADGSRWP+E+
Sbjct: 121 GGQETTPLTAITQLTHHGMVFVPVGYTFGAKMFNMGEVQGGSPYGAGTFAADGSRWPTEM 180
Query: 296 ELEHAFHQGKYFAGIAKKLKGSA 228
ELEHAFHQGKYFAGIAKKLKGSA
Sbjct: 181 ELEHAFHQGKYFAGIAKKLKGSA 203
>sptr|Q9XH74|Q9XH74 Hypothetical 21.7 kDa protein.
Length = 204
Score = 344 bits (882), Expect = 1e-93
Identities = 161/202 (79%), Positives = 185/202 (91%)
Frame = -1
Query: 836 MAVKVYVVYYSMYGHVGKLAEEIQKGASSVEGVEAKTWQVPETLPEEVLGKMGAPPKPDV 657
MA KVY+VYYSMYGHV KLAEEI KGA+SVEGVEAK WQV ETL ++VLGKMGAPPK +V
Sbjct: 1 MATKVYIVYYSMYGHVEKLAEEILKGAASVEGVEAKLWQVAETLQDDVLGKMGAPPKSEV 60
Query: 656 PVITPQELAEADGILFGFPTRFGMMAAQMKAFFDATGGLWREQSLAGKPAGVFFCTGSQG 477
P+I+P +L+EADG+LFGFPTRFGMMAAQ KAFFD+TGGLWR Q+LAGKPAG+F+ TGSQG
Sbjct: 61 PIISPNDLSEADGLLFGFPTRFGMMAAQFKAFFDSTGGLWRTQALAGKPAGIFYSTGSQG 120
Query: 476 GGQETTALTAITQLTHHGMVFVPVGYTFGAKLFGMDQVQGGSPYGAGTFAADGSRWPSEV 297
GGQETTALTAITQL HHGMVFVP+GY+FGA +F M+Q++GGSPYGAGT+A DG+R PSE+
Sbjct: 121 GGQETTALTAITQLVHHGMVFVPIGYSFGAGMFEMEQIKGGSPYGAGTYAGDGTRQPSEL 180
Query: 296 ELEHAFHQGKYFAGIAKKLKGS 231
EL+ AFHQGKYFAGIAKKLKGS
Sbjct: 181 ELQQAFHQGKYFAGIAKKLKGS 202
>sptr|Q8L5Q7|Q8L5Q7 Putative quinone oxidoreductase (EC 1.6.5.5).
Length = 204
Score = 343 bits (881), Expect = 2e-93
Identities = 161/199 (80%), Positives = 181/199 (90%)
Frame = -1
Query: 827 KVYVVYYSMYGHVGKLAEEIQKGASSVEGVEAKTWQVPETLPEEVLGKMGAPPKPDVPVI 648
KVY+VYYS YGHV KLAEEIQKGA+SVEGVEAK WQVPETLPE+VLGKMGAPPK DVP+I
Sbjct: 5 KVYIVYYSTYGHVHKLAEEIQKGAASVEGVEAKLWQVPETLPEDVLGKMGAPPKSDVPII 64
Query: 647 TPQELAEADGILFGFPTRFGMMAAQMKAFFDATGGLWREQSLAGKPAGVFFCTGSQGGGQ 468
TP EL EADG+LFGFPTRFGMMAAQ KAF DATGGLWR Q+LAGKPAG+F+ TGSQGGGQ
Sbjct: 65 TPNELPEADGLLFGFPTRFGMMAAQFKAFMDATGGLWRTQALAGKPAGIFYSTGSQGGGQ 124
Query: 467 ETTALTAITQLTHHGMVFVPVGYTFGAKLFGMDQVQGGSPYGAGTFAADGSRWPSEVELE 288
ETT LT+ITQL HHG++FVP+GYTFGA +F +++V+GGSPYGAGT+A DGSR P+E+EL
Sbjct: 125 ETTPLTSITQLVHHGLIFVPIGYTFGAGMFEIEKVKGGSPYGAGTYAGDGSRQPTELELA 184
Query: 287 HAFHQGKYFAGIAKKLKGS 231
AFHQGKYFAGIAKKLKGS
Sbjct: 185 QAFHQGKYFAGIAKKLKGS 203
>sptr|Q9LSQ5|Q9LSQ5 1,4-benzoquinone reductase-like, Trp repressor
binding protein-like (1,4-benzoquinone reductase-like
protein).
Length = 204
Score = 335 bits (858), Expect = 7e-91
Identities = 159/202 (78%), Positives = 177/202 (87%)
Frame = -1
Query: 836 MAVKVYVVYYSMYGHVGKLAEEIQKGASSVEGVEAKTWQVPETLPEEVLGKMGAPPKPDV 657
MA KVY+VYYSMYGHV KLAEEI+KGA+SVEGVEAK WQVPETL EE L KM APPK +
Sbjct: 1 MATKVYIVYYSMYGHVEKLAEEIRKGAASVEGVEAKLWQVPETLHEEALSKMSAPPKSES 60
Query: 656 PVITPQELAEADGILFGFPTRFGMMAAQMKAFFDATGGLWREQSLAGKPAGVFFCTGSQG 477
P+ITP ELAEADG +FGFPTRFGMMAAQ KAF DATGGLWR Q+LAGKPAG+F+ TGSQG
Sbjct: 61 PIITPNELAEADGFVFGFPTRFGMMAAQFKAFLDATGGLWRAQALAGKPAGIFYSTGSQG 120
Query: 476 GGQETTALTAITQLTHHGMVFVPVGYTFGAKLFGMDQVQGGSPYGAGTFAADGSRWPSEV 297
GGQETTALTAITQL HHGM+FVP+GYTFGA +F M+ V+GGSPYGAGTFA DGSR P+E+
Sbjct: 121 GGQETTALTAITQLVHHGMLFVPIGYTFGAGMFEMENVKGGSPYGAGTFAGDGSRQPTEL 180
Query: 296 ELEHAFHQGKYFAGIAKKLKGS 231
EL+ AFHQG+Y A I KKLKGS
Sbjct: 181 ELQQAFHQGQYIASITKKLKGS 202
>sptrnew|AAQ65137|AAQ65137 At4g27270.
Length = 205
Score = 333 bits (854), Expect = 2e-90
Identities = 158/203 (77%), Positives = 176/203 (86%)
Frame = -1
Query: 836 MAVKVYVVYYSMYGHVGKLAEEIQKGASSVEGVEAKTWQVPETLPEEVLGKMGAPPKPDV 657
MA KVY+VYYSMYGHV KLA+EI+KGA+SV+GVEA WQVPETL E+VL KM APPK D
Sbjct: 1 MATKVYIVYYSMYGHVEKLAQEIRKGAASVDGVEAILWQVPETLQEDVLSKMSAPPKSDA 60
Query: 656 PVITPQELAEADGILFGFPTRFGMMAAQMKAFFDATGGLWREQSLAGKPAGVFFCTGSQG 477
P+ITP ELAEADG +FGFPTRFGMMAAQ KAF DATGGLWR Q LAGKPAG+F+ TGSQG
Sbjct: 61 PIITPNELAEADGFIFGFPTRFGMMAAQFKAFLDATGGLWRTQQLAGKPAGIFYSTGSQG 120
Query: 476 GGQETTALTAITQLTHHGMVFVPVGYTFGAKLFGMDQVQGGSPYGAGTFAADGSRWPSEV 297
GGQETTALTAITQL HHGM+FVP+GYTFGA +F M+ V+GGSPYGAGTFA DGSR P+E+
Sbjct: 121 GGQETTALTAITQLVHHGMIFVPIGYTFGAGMFEMENVKGGSPYGAGTFAGDGSRQPTEL 180
Query: 296 ELEHAFHQGKYFAGIAKKLKGSA 228
EL AFHQGKY A I+KKLKG A
Sbjct: 181 ELGQAFHQGKYIAAISKKLKGPA 203
>sptr|Q9AYU0|Q9AYU0 Quinone-oxidoreductase QR2.
Length = 205
Score = 325 bits (832), Expect = 8e-88
Identities = 153/201 (76%), Positives = 176/201 (87%), Gaps = 1/201 (0%)
Frame = -1
Query: 836 MAVKVYVVYYSMYGHVGKLAEEIQKGASSVEGVEAKTWQVPETLPEEVLGKMGAPPKPDV 657
MA KVY+VYYS YGHV +LA+EI+KGA SV VE K WQVPE L +EVLGKM APPK DV
Sbjct: 1 MATKVYIVYYSTYGHVERLAQEIKKGAESVGNVEVKLWQVPEILSDEVLGKMWAPPKSDV 60
Query: 656 PVITPQELAEADGILFGFPTRFGMMAAQMKAFFDATGGLWREQSLAGKPAGVFFCTGSQG 477
PVITP EL EADGI+FGFPTRFGMMAAQ KAFFD+TGGLW+ Q+LAGKPAG+FF TG+QG
Sbjct: 61 PVITPDELVEADGIIFGFPTRFGMMAAQFKAFFDSTGGLWKTQALAGKPAGIFFSTGTQG 120
Query: 476 GGQETTALTAITQLTHHGMVFVPVGYTFGAKLFGMDQVQGGSPYGAGTFA-ADGSRWPSE 300
GGQETTALTAITQLTHHGM++VP+GYTFGA +F M++++GGSPYGAGTFA ADGSR PS+
Sbjct: 121 GGQETTALTAITQLTHHGMIYVPIGYTFGADMFNMEKIKGGSPYGAGTFAGADGSRQPSD 180
Query: 299 VELEHAFHQGKYFAGIAKKLK 237
+EL+ AFHQG Y AGI KK+K
Sbjct: 181 IELKQAFHQGMYIAGITKKIK 201
>sptr|O81830|O81830 Hypothetical 22.3 kDa protein.
Length = 211
Score = 310 bits (795), Expect = 1e-83
Identities = 152/209 (72%), Positives = 173/209 (82%), Gaps = 6/209 (2%)
Frame = -1
Query: 836 MAVKVYVVY----YSMYGHVG--KLAEEIQKGASSVEGVEAKTWQVPETLPEEVLGKMGA 675
MA KVY+VY + + G + KLA+EI+KGA+SV+GVEA WQVPETL E+VL KM A
Sbjct: 1 MATKVYIVYVKIIFCLSGAIDNLKLAQEIRKGAASVDGVEAILWQVPETLQEDVLSKMSA 60
Query: 674 PPKPDVPVITPQELAEADGILFGFPTRFGMMAAQMKAFFDATGGLWREQSLAGKPAGVFF 495
PPK D P+ITP ELAEADG +FGFPTRFGMMAAQ KAF DATGGLWR Q LAGKPAG+F+
Sbjct: 61 PPKSDAPIITPNELAEADGFIFGFPTRFGMMAAQFKAFLDATGGLWRTQQLAGKPAGIFY 120
Query: 494 CTGSQGGGQETTALTAITQLTHHGMVFVPVGYTFGAKLFGMDQVQGGSPYGAGTFAADGS 315
TGSQGGGQETTALTAITQL HHGM+FVP+GYTFGA +F M+ V+GGSPYGAGTFA DGS
Sbjct: 121 STGSQGGGQETTALTAITQLVHHGMIFVPIGYTFGAGMFEMENVKGGSPYGAGTFAGDGS 180
Query: 314 RWPSEVELEHAFHQGKYFAGIAKKLKGSA 228
R P+E+EL AFHQGKY A I+KKLKG A
Sbjct: 181 RQPTELELGQAFHQGKYIAAISKKLKGPA 209
>sptr|O23807|O23807 LEDI-3 protein.
Length = 201
Score = 295 bits (756), Expect = 5e-79
Identities = 146/201 (72%), Positives = 167/201 (83%), Gaps = 1/201 (0%)
Frame = -1
Query: 836 MAVKVYVVYYSMYGHVGKLAEEIQKGASSVEGVEAKTWQVPETLPEEVLGKMGAPPKPDV 657
MAVK+Y+VYYS YGHV KLAEE++KGA SV+GVE K WQV ETLPE+VLGKM APPK D
Sbjct: 1 MAVKLYIVYYSTYGHVLKLAEEMKKGAESVDGVEVKLWQVAETLPEDVLGKMYAPPKGDA 60
Query: 656 PVITPQELAEADGILFGFPTRFGMMAAQMKAFFDATGGLWREQSLAGKPAGVFFCTGSQG 477
P+ITP ELAEADG LFGFPTRFGMM AQ KAFFD+TGGLW+ L GKPAG F +GSQG
Sbjct: 61 PIITPDELAEADGYLFGFPTRFGMMPAQFKAFFDSTGGLWQSGGLVGKPAGFFISSGSQG 120
Query: 476 GGQETTALTAITQLTHHGMVFVPVGYTFGAKLFGMDQVQGGSPYGAGTFAA-DGSRWPSE 300
GGQETTALTAITQL HHGM++VP G TFGA L ++++GG+ YGAGTFA DG+R P+E
Sbjct: 121 GGQETTALTAITQLVHHGMIYVPTGGTFGAGLH-KNEIRGGTLYGAGTFAGPDGARQPTE 179
Query: 299 VELEHAFHQGKYFAGIAKKLK 237
+ELE AFHQGKY AGI K+LK
Sbjct: 180 LELELAFHQGKYTAGIVKRLK 200
>sptr|Q9LUX9|Q9LUX9 1,4-benzoquinone reductase-like (Putative light
harvesting pigment protein).
Length = 207
Score = 273 bits (698), Expect = 3e-72
Identities = 129/202 (63%), Positives = 165/202 (81%), Gaps = 2/202 (0%)
Frame = -1
Query: 836 MAV-KVYVVYYSMYGHVGKLAEEIQKGASSVEGVEAKTWQVPETLPEEVLGKMGAPPKPD 660
MAV K+Y+VYYS++GHV +A E+ +G +SV VEA WQVPETLPE++L K+ A P+PD
Sbjct: 1 MAVTKIYIVYYSLHGHVETMAREVLRGVNSVPDVEATLWQVPETLPEKILEKVKAVPRPD 60
Query: 659 -VPVITPQELAEADGILFGFPTRFGMMAAQMKAFFDATGGLWREQSLAGKPAGVFFCTGS 483
VP I P++LAEADG +FGFP+RFG+MA+Q+ FFD T LW Q+LAGKPAG+F+ TG
Sbjct: 61 DVPDIRPEQLAEADGFMFGFPSRFGVMASQVMTFFDNTNDLWTTQALAGKPAGIFWSTGF 120
Query: 482 QGGGQETTALTAITQLTHHGMVFVPVGYTFGAKLFGMDQVQGGSPYGAGTFAADGSRWPS 303
GGGQE TALTA+T+L HHGM+FVPVGYTFG ++ M +V+GGSPYG+GT+AADGSR P+
Sbjct: 121 HGGGQELTALTAVTKLAHHGMIFVPVGYTFGKSMYEMGEVKGGSPYGSGTYAADGSREPT 180
Query: 302 EVELEHAFHQGKYFAGIAKKLK 237
E+E++ A + GKYFAGIAKKLK
Sbjct: 181 ELEIQQANYHGKYFAGIAKKLK 202
>sptr|O23207|O23207 Minor allergen.
Length = 273
Score = 270 bits (691), Expect = 2e-71
Identities = 132/198 (66%), Positives = 157/198 (79%), Gaps = 1/198 (0%)
Frame = -1
Query: 830 VKVYVVYYSMYGHVGKLAEEIQKGASSVEGVEAKTWQVPETLPEEVLGKMGAPPKP-DVP 654
+K++VV+YSMYGHV LA+ ++KG SVEGVEA ++VPETL +EV+ +M AP K ++P
Sbjct: 73 LKIFVVFYSMYGHVESLAKRMKKGVDSVEGVEATLYRVPETLSQEVVEQMKAPVKDLEIP 132
Query: 653 VITPQELAEADGILFGFPTRFGMMAAQMKAFFDATGGLWREQSLAGKPAGVFFCTGSQGG 474
IT EL ADG LFGFPTR+G MAAQMKAFFD+TG LW+EQSLAGKPAG F TG+QGG
Sbjct: 133 EITAAELTAADGFLFGFPTRYGCMAAQMKAFFDSTGSLWKEQSLAGKPAGFFVSTGTQGG 192
Query: 473 GQETTALTAITQLTHHGMVFVPVGYTFGAKLFGMDQVQGGSPYGAGTFAADGSRWPSEVE 294
GQETTA TAITQL HHGM+FVP+GYTFGA +F MD ++GGSPYGAG FA DGSR +E E
Sbjct: 193 GQETTAWTAITQLVHHGMLFVPIGYTFGAGMFKMDSIRGGSPYGAGVFAGDGSREATETE 252
Query: 293 LEHAFHQGKYFAGIAKKL 240
L A HQG Y A I K+L
Sbjct: 253 LALAEHQGNYMAAIVKRL 270
>sptr|Q8LB59|Q8LB59 Minor allergen.
Length = 273
Score = 270 bits (691), Expect = 2e-71
Identities = 132/198 (66%), Positives = 157/198 (79%), Gaps = 1/198 (0%)
Frame = -1
Query: 830 VKVYVVYYSMYGHVGKLAEEIQKGASSVEGVEAKTWQVPETLPEEVLGKMGAPPKP-DVP 654
+K++VV+YSMYGHV LA+ ++KG SVEGVEA ++VPETL +EV+ +M AP K ++P
Sbjct: 73 LKIFVVFYSMYGHVESLAKRMKKGVDSVEGVEATLYRVPETLSQEVVEQMKAPVKDLEIP 132
Query: 653 VITPQELAEADGILFGFPTRFGMMAAQMKAFFDATGGLWREQSLAGKPAGVFFCTGSQGG 474
IT EL ADG LFGFPTR+G MAAQMKAFFD+TG LW+EQSLAGKPAG F TG+QGG
Sbjct: 133 EITAAELTAADGFLFGFPTRYGCMAAQMKAFFDSTGSLWKEQSLAGKPAGFFVSTGTQGG 192
Query: 473 GQETTALTAITQLTHHGMVFVPVGYTFGAKLFGMDQVQGGSPYGAGTFAADGSRWPSEVE 294
GQETTA TAITQL HHGM+FVP+GYTFGA +F MD ++GGSPYGAG FA DGSR +E E
Sbjct: 193 GQETTAWTAITQLVHHGMLFVPIGYTFGAGMFKMDSIRGGSPYGAGVFAGDGSREATETE 252
Query: 293 LEHAFHQGKYFAGIAKKL 240
L A HQG Y A I K+L
Sbjct: 253 LALAEHQGNYMAAIVKRL 270
>sptr|Q8W305|Q8W305 Putative reductase.
Length = 252
Score = 238 bits (607), Expect = 9e-62
Identities = 123/219 (56%), Positives = 150/219 (68%), Gaps = 11/219 (5%)
Frame = -1
Query: 863 RNSDSEPSPMA----VKVYVVYYSMYGHVGKLAEEIQKGASSVEGVEAKTWQVPETLPEE 696
R S P+P A V++YVV+YSMYGHV LA + +G SV G A ++VPETLP
Sbjct: 29 RERTSIPAPAAAPRPVRIYVVFYSMYGHVRLLARAVARGVGSVPGARAILFRVPETLPPA 88
Query: 695 VLGKM-------GAPPKPDVPVITPQELAEADGILFGFPTRFGMMAAQMKAFFDATGGLW 537
VL +M G + +PV+ P L +ADG LFGFP RFG M AQM+AFFD+T L
Sbjct: 89 VLARMEADDGGGGGDGEDVIPVVDPDGLPDADGFLFGFPARFGAMPAQMQAFFDSTVPLC 148
Query: 536 REQSLAGKPAGVFFCTGSQGGGQETTALTAITQLTHHGMVFVPVGYTFGAKLFGMDQVQG 357
R Q LAGKPAG+F TG+Q GGQETTA TAITQL HHGM+FVP+GYTFG + M +++G
Sbjct: 149 RHQRLAGKPAGLFVSTGTQAGGQETTAWTAITQLAHHGMLFVPIGYTFGEGMLEMGELRG 208
Query: 356 GSPYGAGTFAADGSRWPSEVELEHAFHQGKYFAGIAKKL 240
GSPYGAG F+ DGSR PSE+EL A H GKY A + KK+
Sbjct: 209 GSPYGAGVFSGDGSRPPSELELALAEHHGKYMATLVKKM 247
>sptr|Q9Y763|Q9Y763 1,4-benzoquinone reductase.
Length = 201
Score = 228 bits (581), Expect = 1e-58
Identities = 118/198 (59%), Positives = 145/198 (73%), Gaps = 2/198 (1%)
Frame = -1
Query: 827 KVYVVYYSMYGHVGKLAEEIQKGASSVEGVEAKTWQVPETLPEEVLGKMGAPPKPDVPVI 648
KV ++ YSMYGH+ KLAE + G G A +Q+PETLPEEVL KM APPKP+ PVI
Sbjct: 3 KVAIIIYSMYGHIAKLAEAEKAGIEEAGG-SATIYQIPETLPEEVLAKMHAPPKPEYPVI 61
Query: 647 TPQELAEADGILFGFPTRFGMMAAQMKAFFDATGGLWREQSLAGKPAGVFFCTGSQGGGQ 468
TP++L E D +FG PTR+G Q KAF+DATGGLW + +LAGK A VF TG+ GGGQ
Sbjct: 62 TPEKLPEFDAFVFGIPTRYGNFPGQWKAFWDATGGLWAQGALAGKYASVFVSTGTPGGGQ 121
Query: 467 ETTALTAITQLTHHGMVFVPVGY-TFGAKLFGMDQVQGGSPYGAGTFA-ADGSRWPSEVE 294
E+T L +I+ LTHHG+VFVP+GY T A+L + +V+GGSP+GAGTFA ADGSR PS +E
Sbjct: 122 ESTVLNSISTLTHHGIVFVPLGYSTTFAQLANLSEVRGGSPWGAGTFAGADGSRSPSALE 181
Query: 293 LEHAFHQGKYFAGIAKKL 240
LE A QGKYF I KK+
Sbjct: 182 LELATAQGKYFWNIIKKV 199
>sptr|Q941C6|Q941C6 C7A10_610/C7A10_610 (At4g36690/C7A10_610).
Length = 152
Score = 212 bits (539), Expect = 7e-54
Identities = 104/149 (69%), Positives = 117/149 (78%), Gaps = 1/149 (0%)
Frame = -1
Query: 683 MGAPPKP-DVPVITPQELAEADGILFGFPTRFGMMAAQMKAFFDATGGLWREQSLAGKPA 507
M AP K ++P IT EL ADG LFGFPTR+G MAAQMKAFFD+TG LW+EQSLAGKPA
Sbjct: 1 MKAPVKDLEIPEITAAELTAADGFLFGFPTRYGCMAAQMKAFFDSTGSLWKEQSLAGKPA 60
Query: 506 GVFFCTGSQGGGQETTALTAITQLTHHGMVFVPVGYTFGAKLFGMDQVQGGSPYGAGTFA 327
G F TG+QGGGQETTA TAITQL HHGM+FVP+GYTFGA +F MD ++GGSPYGAG FA
Sbjct: 61 GFFVSTGTQGGGQETTAWTAITQLVHHGMLFVPIGYTFGAGMFKMDSIRGGSPYGAGVFA 120
Query: 326 ADGSRWPSEVELEHAFHQGKYFAGIAKKL 240
DGSR +E EL A HQG Y A I K+L
Sbjct: 121 GDGSREATETELALAEHQGNYMAAIVKRL 149
>sw|Q8Z7N9|WRBA_SALTI Flavoprotein wrbA (Trp repressor binding
protein).
Length = 197
Score = 201 bits (512), Expect = 1e-50
Identities = 105/199 (52%), Positives = 134/199 (67%), Gaps = 1/199 (0%)
Frame = -1
Query: 827 KVYVVYYSMYGHVGKLAEEIQKGASSVEGVEAKTWQVPETLPEEVLGKMGAPPKPDVPVI 648
K+ V+YYSMYGH+ +A + +GA V+G E +VPET+P E+ K G + + PV
Sbjct: 2 KILVLYYSMYGHIETMAHAVAEGAKKVDGAEVIIKRVPETMPPEIFAKAGGKTQ-NAPVA 60
Query: 647 TPQELAEADGILFGFPTRFGMMAAQMKAFFDATGGLWREQSLAGKPAGVFFCTGSQGGGQ 468
TPQELA+ D I+FG PTRFG M+ QM+ F D TGGLW +L GK GVF TG+ GGGQ
Sbjct: 61 TPQELADYDAIIFGTPTRFGNMSGQMRTFLDQTGGLWASGALYGKLGGVFSSTGT-GGGQ 119
Query: 467 ETTALTAITQLTHHGMVFVPVGYTFGAKLFGMDQVQGGSPYGAGTFA-ADGSRWPSEVEL 291
E T + T L HHGMV VP+GY+ +LF + QV+GG+PYGA T A DGSR PS+ EL
Sbjct: 120 EQTITSTWTTLAHHGMVIVPIGYS-AQELFDVSQVRGGTPYGATTIAGGDGSRQPSQEEL 178
Query: 290 EHAFHQGKYFAGIAKKLKG 234
A +QG+Y AG+A KL G
Sbjct: 179 SIARYQGEYVAGLAVKLNG 197
>sw|Q8ZQ40|WRBA_SALTY Flavoprotein wrbA (Trp repressor binding
protein).
Length = 197
Score = 200 bits (508), Expect = 3e-50
Identities = 105/199 (52%), Positives = 132/199 (66%), Gaps = 1/199 (0%)
Frame = -1
Query: 827 KVYVVYYSMYGHVGKLAEEIQKGASSVEGVEAKTWQVPETLPEEVLGKMGAPPKPDVPVI 648
K+ V+YYSMYGH+ +A + +GA V+G E +VPET+P E+ K G + + PV
Sbjct: 2 KILVLYYSMYGHIETMAHAVAEGAKKVDGAEVIIKRVPETMPPEIFAKAGGKTQ-NAPVA 60
Query: 647 TPQELAEADGILFGFPTRFGMMAAQMKAFFDATGGLWREQSLAGKPAGVFFCTGSQGGGQ 468
TPQELA+ D I+FG PTRFG M+ QM+ F D TGGLW SL GK VF TG+ GGGQ
Sbjct: 61 TPQELADYDAIIFGTPTRFGNMSGQMRTFLDQTGGLWASGSLYGKLGSVFSSTGT-GGGQ 119
Query: 467 ETTALTAITQLTHHGMVFVPVGYTFGAKLFGMDQVQGGSPYGAGTFA-ADGSRWPSEVEL 291
E T + T L HHGMV VP+GY +LF + QV+GG+PYGA T A DGSR PS+ EL
Sbjct: 120 EQTITSTWTTLAHHGMVIVPIGYA-AQELFDVSQVRGGTPYGATTIAGGDGSRQPSQEEL 178
Query: 290 EHAFHQGKYFAGIAKKLKG 234
A +QG+Y AG+A KL G
Sbjct: 179 SIARYQGEYVAGLAVKLNG 197
>sw|P30849|WRBA_ECOLI Flavoprotein wrbA (Trp repressor binding
protein).
Length = 197
Score = 199 bits (507), Expect = 4e-50
Identities = 106/199 (53%), Positives = 133/199 (66%), Gaps = 1/199 (0%)
Frame = -1
Query: 827 KVYVVYYSMYGHVGKLAEEIQKGASSVEGVEAKTWQVPETLPEEVLGKMGAPPKPDVPVI 648
KV V+YYSMYGH+ +A + +GAS V+G E +VPET+P ++ K G + PV
Sbjct: 2 KVLVLYYSMYGHIETMARAVAEGASKVDGAEVVVKRVPETMPPQLFEKAGGKTQT-APVA 60
Query: 647 TPQELAEADGILFGFPTRFGMMAAQMKAFFDATGGLWREQSLAGKPAGVFFCTGSQGGGQ 468
TPQELA+ D I+FG PTRFG M+ QM+ F D TGGLW +L GK A VF TG+ GGGQ
Sbjct: 61 TPQELADYDAIIFGTPTRFGNMSGQMRTFLDQTGGLWASGALYGKLASVFSSTGT-GGGQ 119
Query: 467 ETTALTAITQLTHHGMVFVPVGYTFGAKLFGMDQVQGGSPYGAGTFA-ADGSRWPSEVEL 291
E T + T L HHGMV VP+GY +LF + QV+GG+PYGA T A DGSR PS+ EL
Sbjct: 120 EQTITSTWTTLAHHGMVIVPIGYA-AQELFDVSQVRGGTPYGATTIAGGDGSRQPSQEEL 178
Query: 290 EHAFHQGKYFAGIAKKLKG 234
A +QG+Y AG+A KL G
Sbjct: 179 SIARYQGEYVAGLAVKLNG 197
>sw|P42058|ALA7_ALTAL Minor allergen Alt a 7 (Alt a VII).
Length = 204
Score = 198 bits (503), Expect = 1e-49
Identities = 110/202 (54%), Positives = 141/202 (69%), Gaps = 5/202 (2%)
Frame = -1
Query: 836 MAVKVYVVYYSMYGHVGKLAEEIQKGASSVEGVEAKTWQVPETLPEEVLGKMGAPPKPD- 660
MA K+ +VYYSMYGH+ K+A+ KG G +AK +QV ETLP+EVL KM APPK
Sbjct: 1 MAPKIAIVYYSMYGHIKKMADAELKGIQEAGG-DAKLFQVAETLPQEVLDKMYAPPKDSS 59
Query: 659 VPVIT-PQELAEADGILFGFPTRFGMMAAQMKAFFDATGGLWREQSLAGKPAGVFFCTGS 483
VPV+ P L E DGILFG PTR+G AQ K F+D TG W++ + GK AGVF TG+
Sbjct: 60 VPVLEDPAVLEEFDGILFGIPTRYGNFPAQFKTFWDKTGKQWQQGAFWGKYAGVFVSTGT 119
Query: 482 QGGGQETTALTAITQLTHHGMVFVPVGY-TFGAKLFGMDQVQGGSPYGAGTFAA-DGSRW 309
GGGQETTA+T+++ L HG ++VP+GY T + L +D+V GGSP+GAGTF+A DGSR
Sbjct: 120 LGGGQETTAITSMSTLVDHGFIYVPLGYKTAFSMLANLDEVHGGSPWGAGTFSAGDGSRQ 179
Query: 308 PSEVELEHAFHQGK-YFAGIAK 246
PSE+EL A QGK ++ +AK
Sbjct: 180 PSELELNIAQAQGKAFYEAVAK 201
>sptrnew|EAA36394|EAA36394 Hypothetical protein.
Length = 244
Score = 196 bits (498), Expect = 4e-49
Identities = 110/209 (52%), Positives = 137/209 (65%), Gaps = 7/209 (3%)
Frame = -1
Query: 836 MAVKVYVVYYSMYGHVGKLAEEIQKGASSVEGVEAKTWQVPETLPEEVLGKMGAPPKP-D 660
MA K+ +VYYSMYGH+ +LAE + G G A +QVPETL +EVL KM APPKP D
Sbjct: 1 MAPKIAIVYYSMYGHIRQLAEAAKAGIEKAGGT-ADLYQVPETLSDEVLAKMYAPPKPTD 59
Query: 659 VPVIT-PQELAEADGILFGFPTRFGMMAAQMKAFFDATGGLWREQSLAGKPAGVFFCTGS 483
+PVI P L E DG LFG PTR+G AQ +AF+D TGGLW L GK AG+F T
Sbjct: 60 IPVIEDPAILKEYDGFLFGIPTRYGNFPAQWRAFWDKTGGLWATGGLYGKAAGLFISTAG 119
Query: 482 QGGGQETTALTAITQLTHHGMVFVPVGYTFGAKLFG----MDQVQGGSPYGAGTFA-ADG 318
GGGQE+TA+ A++ L HHG+++VP+GY AK+FG + V GGSP+G+GT + DG
Sbjct: 120 LGGGQESTAIAAMSTLAHHGIIYVPLGY---AKVFGELSDLSAVHGGSPWGSGTLSGGDG 176
Query: 317 SRWPSEVELEHAFHQGKYFAGIAKKLKGS 231
SR PSE EL+ A QG+ F KL S
Sbjct: 177 SRQPSESELKVAGIQGEEFYNTLSKLTAS 205
>sw|Q8X4B4|WRBA_ECO57 Flavoprotein wrbA (Trp repressor binding
protein).
Length = 197
Score = 196 bits (497), Expect = 5e-49
Identities = 105/199 (52%), Positives = 131/199 (65%), Gaps = 1/199 (0%)
Frame = -1
Query: 827 KVYVVYYSMYGHVGKLAEEIQKGASSVEGVEAKTWQVPETLPEEVLGKMGAPPKPDVPVI 648
KV V+YYSMYGH+ +A + +GAS V+G E +VPET+ ++ K G + PV
Sbjct: 2 KVLVLYYSMYGHIETMARAVAEGASKVDGAEVVVKRVPETMSPQLFEKAGGKTQT-APVA 60
Query: 647 TPQELAEADGILFGFPTRFGMMAAQMKAFFDATGGLWREQSLAGKPAGVFFCTGSQGGGQ 468
TPQELA D I+FG PTRFG M+ QM+ F D TGGLW +L GK A VF TG+ GGGQ
Sbjct: 61 TPQELANYDAIIFGTPTRFGNMSGQMRTFLDQTGGLWASGALYGKLASVFSSTGT-GGGQ 119
Query: 467 ETTALTAITQLTHHGMVFVPVGYTFGAKLFGMDQVQGGSPYGAGTFA-ADGSRWPSEVEL 291
E T + T L HHGMV VP+GY +LF + QV+GG+PYGA T A DGSR PS+ EL
Sbjct: 120 EQTITSTWTTLAHHGMVIVPIGYA-AQELFDVSQVRGGTPYGATTIAGGDGSRQPSQEEL 178
Query: 290 EHAFHQGKYFAGIAKKLKG 234
A +QG+Y AG+A KL G
Sbjct: 179 SIARYQGEYVAGLAVKLNG 197
>sptr|Q8X172|Q8X172 NADH:quinone oxidoreductase.
Length = 257
Score = 196 bits (497), Expect = 5e-49
Identities = 101/199 (50%), Positives = 135/199 (67%), Gaps = 2/199 (1%)
Frame = -1
Query: 827 KVYVVYYSMYGHVGKLAEEIQKGASSVEGVEAKTWQVPETLPEEVLGKMGAPPKPDVPVI 648
++ +V Y+MYGHV KLAE I+ G G A +QV ETL E+L + APPKPD PV+
Sbjct: 59 RLAIVIYTMYGHVAKLAEAIKSGIEGAGG-NASIFQVAETLSPEILNLVKAPPKPDYPVM 117
Query: 647 TPQELAEADGILFGFPTRFGMMAAQMKAFFDATGGLWREQSLAGKPAGVFFCTGSQGGGQ 468
P +L DG LFG PTR+G Q KAF+D+TG LW +L GK AG+F TGS GGGQ
Sbjct: 118 DPLDLKNYDGFLFGIPTRYGNFPVQWKAFWDSTGPLWASTALCGKYAGLFVSTGSPGGGQ 177
Query: 467 ETTALTAITQLTHHGMVFVPVGYTFG-AKLFGMDQVQGGSPYGAGTFA-ADGSRWPSEVE 294
E+T + A++ L HHG+++VP+GY + A+L + +V+GGSP+GAGTFA +DGSR P+ +E
Sbjct: 178 ESTLMAAMSTLVHHGVIYVPLGYKYTFAQLANLTEVRGGSPWGAGTFANSDGSRQPTPLE 237
Query: 293 LEHAFHQGKYFAGIAKKLK 237
LE A QGK F ++K
Sbjct: 238 LEIANLQGKSFYEYVARVK 256
>sw|Q8ZF61|WRBA_YERPE Flavoprotein wrbA (Trp repressor binding
protein).
Length = 199
Score = 194 bits (493), Expect = 2e-48
Identities = 106/200 (53%), Positives = 127/200 (63%), Gaps = 2/200 (1%)
Frame = -1
Query: 827 KVYVVYYSMYGHVGKLAEEIQKGASSVEGVEAKTWQVPETLPEEVLGKMGAPPKPDVPVI 648
K+ V+YYSMYGH+ LA I +GA V GV+ +VPET+P E K G PV
Sbjct: 3 KILVLYYSMYGHIETLAGAIAEGARKVSGVDVTIKRVPETMPAEAFAKAGGKTNQQAPVA 62
Query: 647 TPQELAEADGILFGFPTRFGMMAAQMKAFFDATGGLWREQSLAGKPAGVFFCTGSQGGGQ 468
TP ELA+ DGI+FG PTRFG M+ QM+ F D TGGLW +L GK A VF TG+ GGGQ
Sbjct: 63 TPHELADYDGIIFGTPTRFGNMSGQMRTFLDQTGGLWASGALYGKVASVFASTGT-GGGQ 121
Query: 467 ETTALTAITQLTHHGMVFVPVGYTFGAK-LFGMDQVQGGSPYGAGTFA-ADGSRWPSEVE 294
E T + T L HHG + VP+GY GAK LF + Q +GG+PYGA T A DGSR PS E
Sbjct: 122 EHTITSTWTTLAHHGFIIVPIGY--GAKELFDVSQTRGGTPYGATTIAGGDGSRQPSAEE 179
Query: 293 LEHAFHQGKYFAGIAKKLKG 234
L A QG++ A I KLKG
Sbjct: 180 LAIARFQGEHVAKITAKLKG 199
>sptr|Q8X194|Q8X194 Y20 protein.
Length = 203
Score = 193 bits (490), Expect = 3e-48
Identities = 101/201 (50%), Positives = 133/201 (66%), Gaps = 2/201 (0%)
Frame = -1
Query: 836 MAVKVYVVYYSMYGHVGKLAEEIQKGASSVEGVEAKTWQVPETLPEEVLGKMGAPPKPDV 657
MA K+ +V+YS+YGH+ KLAE +KG + G A +Q+ ETLP+EVL KM AP K
Sbjct: 1 MAPKIAIVFYSLYGHIQKLAEAEKKGIEAAGGT-ADIYQIAETLPQEVLDKMHAPGKSSY 59
Query: 656 PVITPQELAEADGILFGFPTRFGMMAAQMKAFFDATGGLWREQSLAGKPAGVFFCTGSQG 477
PV P L D LFG PTR+G Q KAF+D TGG+W GK AG+F TG+ G
Sbjct: 60 PVAEPATLLNYDAFLFGIPTRYGNFPGQWKAFWDKTGGIWSTGGFWGKYAGLFVSTGTPG 119
Query: 476 GGQETTALTAITQLTHHGMVFVPVGY-TFGAKLFGMDQVQGGSPYGAGTFA-ADGSRWPS 303
GGQE+T + A++ L HHG+++VP+GY T L +++V+GGSP+GAGT+A ADGSR PS
Sbjct: 120 GGQESTNIAAMSTLAHHGIIYVPLGYKTTFPILANLNEVRGGSPWGAGTYAGADGSRQPS 179
Query: 302 EVELEHAFHQGKYFAGIAKKL 240
+EL+ A QGK F G K+
Sbjct: 180 ALELQLAEEQGKAFYGAVSKV 200
>sw|P25349|YCP4_YEAST Hypothetical 26.4 kDa protein in CDC10-CIT2
intergenic region.
Length = 247
Score = 192 bits (487), Expect = 8e-48
Identities = 103/200 (51%), Positives = 134/200 (67%), Gaps = 3/200 (1%)
Frame = -1
Query: 830 VKVYVVYYSMYGHVGKLAEEIQKGASSVEGVEAKTWQVPETLPEEVLGKMGAPPKP-DVP 654
VK+ ++ YS YGH+ LA+ ++KG + G +A ++V ETLP+EVL KM AP KP D+P
Sbjct: 2 VKIAIITYSTYGHIDVLAQAVKKGVEAAGG-KADIYRVEETLPDEVLTKMNAPQKPEDIP 60
Query: 653 VITPQELAEADGILFGFPTRFGMMAAQMKAFFDATGGLWREQSLAGKPAGVFFCTGSQGG 474
V T + L E D LFG PTRFG + AQ AF+D TGGLW + SL GK AG+F T S GG
Sbjct: 61 VATEKTLLEYDAFLFGVPTRFGNLPAQWSAFWDKTGGLWAKGSLNGKAAGIFVSTSSYGG 120
Query: 473 GQETTALTAITQLTHHGMVFVPVGYTFG-AKLFGMDQVQGGSPYGAGTFAA-DGSRWPSE 300
GQE+T ++ L HHG++F+P+GY A+L +++V GGSP+GAGT A DGSR S
Sbjct: 121 GQESTVKACLSYLAHHGIIFLPLGYKNSFAELASIEEVHGGSPWGAGTLAGPDGSRTASP 180
Query: 299 VELEHAFHQGKYFAGIAKKL 240
+EL A QGK F AKKL
Sbjct: 181 LELRIAEIQGKTFYETAKKL 200
>sw|P58796|WRBA_METAC Flavoprotein wrbA.
Length = 209
Score = 181 bits (459), Expect = 1e-44
Identities = 102/208 (49%), Positives = 130/208 (62%), Gaps = 9/208 (4%)
Frame = -1
Query: 830 VKVYVVYYSMYGHVGKLAEEIQKGASSVEGVEAKTWQVPETLPEEVLGKMGAPPKPD--- 660
VKV +++YSMYGHV ++AE + GA VEG E +QVPETLPEEVL KMGA
Sbjct: 2 VKVNIIFYSMYGHVYRMAEAVAAGAREVEGAEVGIYQVPETLPEEVLEKMGAIETKKLFA 61
Query: 659 -VPVIT----PQELAEADGILFGFPTRFGMMAAQMKAFFDATGGLWREQSLAGKPAGVFF 495
+PV+T + LA AD ++FG PTR+G M AQM+A D GGLW + GK VF
Sbjct: 62 HIPVLTREMNEEVLAGADALIFGTPTRYGNMTAQMRAVLDGLGGLWNRDAFVGKVGSVFT 121
Query: 494 CTGSQGGGQETTALTAITQLTHHGMVFVPVGYTFGAKLFGMDQVQGGSPYGAGTFA-ADG 318
+G+Q GGQE+T LT L H GM+ V + Y+ + MD++ GGSPYG T A DG
Sbjct: 122 SSGTQHGGQESTILTFHVTLLHLGMILVGLPYS-EKRQTRMDEITGGSPYGVSTIAGGDG 180
Query: 317 SRWPSEVELEHAFHQGKYFAGIAKKLKG 234
SR PSE EL A +QG++ IAKK+ G
Sbjct: 181 SRQPSENELAMARYQGRHVTLIAKKIAG 208
>sw|P30821|P25_SCHPO P25 protein (Brefeldin A resistance protein).
Length = 202
Score = 180 bits (456), Expect = 3e-44
Identities = 99/191 (51%), Positives = 129/191 (67%), Gaps = 3/191 (1%)
Frame = -1
Query: 824 VYVVYYSMYGHVGKLAEEIQKGASSVEGVEAKTWQVPETLPEEVLGKMGAPPKPDVPVIT 645
V +V YS YGHV KLAE + G G +A +Q PETL E+L KM A PKP+ PV+T
Sbjct: 7 VAIVIYSTYGHVVKLAEAEKAGIEKAGG-KAVIYQFPETLSPEILEKMHAAPKPNYPVVT 65
Query: 644 PQELAEADGILFGFPTRFGMMAAQMKAFFDATGGLWREQSLAGKPAGVFFCTGSQGGGQE 465
L + D LFG+PTR+G AQ + F+D+TGGLW + +L GK G FF TG+ GGGQE
Sbjct: 66 LDVLTQYDAFLFGYPTRYGTPPAQFRTFWDSTGGLWVQGALHGKYFGQFFSTGTLGGGQE 125
Query: 464 TTALTAITQLTHHGMVFVPVGY--TFGAKLFGMDQVQGGSPYGAGTFA-ADGSRWPSEVE 294
+TALTA+T HHGM+FVP+GY TF + + ++ + GGS +GAG++A ADGSR S+ E
Sbjct: 126 STALTAMTSFVHHGMIFVPLGYKNTF-SLMANVESIHGGSSWGAGSYAGADGSRNVSDDE 184
Query: 293 LEHAFHQGKYF 261
LE A QG+ F
Sbjct: 185 LEIARIQGETF 195
>sw|Q12335|PST2_YEAST Protoplast secreted protein 2 precursor.
Length = 198
Score = 179 bits (453), Expect = 7e-44
Identities = 96/191 (50%), Positives = 124/191 (64%), Gaps = 2/191 (1%)
Frame = -1
Query: 827 KVYVVYYSMYGHVGKLAEEIQKGASSVEGVEAKTWQVPETLPEEVLGKMGAPPKPDVPVI 648
+V ++ Y++YGHV AE +KG + G A +QV ETL EV+ +G PKPD P+
Sbjct: 3 RVAIIIYTLYGHVAATAEAEKKGIEAAGG-SADIYQVEETLSPEVVKALGGAPKPDYPIA 61
Query: 647 TPQELAEADGILFGFPTRFGMMAAQMKAFFDATGGLWREQSLAGKPAGVFFCTGSQGGGQ 468
T L E D LFG PTRFG AQ KAF+D TGGLW + +L GK AG F TG+ GGG
Sbjct: 62 TQDTLTEYDAFLFGIPTRFGNFPAQWKAFWDRTGGLWAKGALHGKVAGCFVSTGT-GGGN 120
Query: 467 ETTALTAITQLTHHGMVFVPVGY-TFGAKLFGMDQVQGGSPYGAGTFA-ADGSRWPSEVE 294
E T + +++ L HHG++FVP+GY A+L MD+V GGSP+GAGT A +DGSR PS +E
Sbjct: 121 EATIMNSLSTLAHHGIIFVPLGYKNVFAELTNMDEVHGGSPWGAGTIAGSDGSRSPSALE 180
Query: 293 LEHAFHQGKYF 261
L+ QGK F
Sbjct: 181 LQVHEIQGKTF 191
>sw|Q8PUV4|WRBA_METMA Flavoprotein wrbA.
Length = 209
Score = 174 bits (441), Expect = 2e-42
Identities = 98/208 (47%), Positives = 130/208 (62%), Gaps = 9/208 (4%)
Frame = -1
Query: 830 VKVYVVYYSMYGHVGKLAEEIQKGASSVEGVEAKTWQVPETLPEEVLGKMGAPPKPD--- 660
VKV ++++S++ H+ ++AE + GA VEG E +QVPETLPE+VL KMGA
Sbjct: 2 VKVNIIFHSVHAHIYRMAEAVAAGAREVEGAEVGIYQVPETLPEDVLEKMGAIETKKLFA 61
Query: 659 -VPVIT----PQELAEADGILFGFPTRFGMMAAQMKAFFDATGGLWREQSLAGKPAGVFF 495
+PV+T LA AD ++FG PTR+GMM AQM+A D G LW E + GK VF
Sbjct: 62 HIPVVTRDMYEDVLAGADALIFGTPTRYGMMTAQMRAVLDGLGKLWSEDAFVGKVGSVFT 121
Query: 494 CTGSQGGGQETTALTAITQLTHHGMVFVPVGYTFGAKLFGMDQVQGGSPYGAGTFA-ADG 318
+G+Q GGQE+T L+ L H GMV V + Y + M+++ GGSPYGA T A DG
Sbjct: 122 SSGTQHGGQESTILSFHVTLLHLGMVIVGLPYAEKRQTI-MNEITGGSPYGASTIAGGDG 180
Query: 317 SRWPSEVELEHAFHQGKYFAGIAKKLKG 234
SR PSE ELE A +QG++ IAKK+ G
Sbjct: 181 SRQPSENELEMARYQGRHVTQIAKKIAG 208
>sw|P42059|CLH5_CLAHE Minor allergen Cla h 5 (Cla h V).
Length = 204
Score = 165 bits (417), Expect = 1e-39
Identities = 93/201 (46%), Positives = 127/201 (63%), Gaps = 4/201 (1%)
Frame = -1
Query: 836 MAVKVYVVYYSMYGHVGKLAEEIQKGASSVEGVEAKTWQVPETLPEEVLGKMGAPPKPD- 660
MA K+ +++YS +GHV LAE KG G ++VPETL +EVL KM APPK D
Sbjct: 1 MAPKIAIIFYSTWGHVQTLAEAEAKGIREAGG-SVDLYRVPETLTQEVLTKMHAPPKDDS 59
Query: 659 VPVIT-PQELAEADGILFGFPTRFGMMAAQMKAFFDATGGLWREQSLAGKPAGVFFCTGS 483
+P IT P L + D G PTR+G AQ + F+D TGG W+ + GK AG+F TG+
Sbjct: 60 IPEITDPFILEQYDRFPHGHPTRYGNFPAQWRTFWDRTGGQWQTGAFWGKYAGLFISTGT 119
Query: 482 QGGGQETTALTAITQLTHHGMVFVPVGYTFGAKLFG-MDQVQGGSPYGAGTFA-ADGSRW 309
QGGGQE+TAL A++ L+HHG+++VP+GY L G +V+G + +GAGTF+ DGSR
Sbjct: 120 QGGGQESTALAAMSTLSHHGIIYVPLGYKTTFHLLGDNSEVRGAAVWGAGTFSGGDGSRQ 179
Query: 308 PSEVELEHAFHQGKYFAGIAK 246
PS+ ELE ++ +AK
Sbjct: 180 PSQKELELTAQGKAFYEAVAK 200
>sw|Q930L2|WRB2_RHIME Flavoprotein wrbA2.
Length = 202
Score = 162 bits (411), Expect = 5e-39
Identities = 91/197 (46%), Positives = 118/197 (59%), Gaps = 1/197 (0%)
Frame = -1
Query: 827 KVYVVYYSMYGHVGKLAEEIQKGASSVEGVEAKTWQVPETLPEEVLGKMGAPPKPDVPVI 648
+V V+YYS YGH+ +A + +GA S G E +VPET+P EV K PV
Sbjct: 3 RVLVLYYSSYGHIETMAGAVAEGARST-GAEVTIKRVPETVPIEVADKAHFKLNQAAPVA 61
Query: 647 TPQELAEADGILFGFPTRFGMMAAQMKAFFDATGGLWREQSLAGKPAGVFFCTGSQGGGQ 468
T ELA+ D I+ G TRFG M++QM F D GGLW +L GK G F TG+Q GGQ
Sbjct: 62 TVAELADYDAIIVGTGTRFGRMSSQMAVFLDQAGGLWARGALNGKVGGAFVSTGTQHGGQ 121
Query: 467 ETTALTAITQLTHHGMVFVPVGYTFGAKLFGMDQVQGGSPYGAGTFA-ADGSRWPSEVEL 291
ETT + IT L H GMV V + Y+ ++ +D++ GG+PYGA T A DGSR PS+++L
Sbjct: 122 ETTLFSIITNLMHFGMVIVGLPYSHQGQM-SVDEIVGGAPYGATTVAGGDGSRQPSQIDL 180
Query: 290 EHAFHQGKYFAGIAKKL 240
AFHQG+ A A L
Sbjct: 181 AGAFHQGEIVARTAAAL 197
Database: /db/trembl-ebi/tmp/swall
Posted date: Sep 26, 2003 8:29 PM
Number of letters in database: 406,886,216
Number of sequences in database: 1,272,877
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 734,574,546
Number of Sequences: 1272877
Number of extensions: 16781940
Number of successful extensions: 74319
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 70006
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 74035
length of database: 406,886,216
effective HSP length: 123
effective length of database: 250,322,345
effective search space used: 49313501965
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

