BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= 2192593.2.1
(800 letters)
Database: /db/uniprot/tmp/swall
1,395,590 sequences; 442,889,342 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
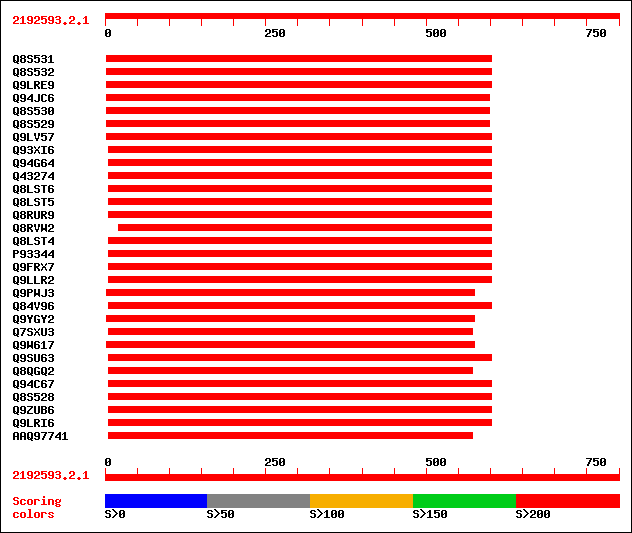
sptr|Q8S531|Q8S531 Cytosolic aldehyde dehydrogenase RF2C. 409 e-113
sptr|Q8S532|Q8S532 Cytosolic aldehyde dehydrogenase RF2C. 402 e-111
sptr|Q9LRE9|Q9LRE9 Cytosolic aldehyde dehydrogenase. 365 e-100
sptr|Q94JC6|Q94JC6 Putative cytosolic aldehyde dehydrogenase. 315 4e-85
sptr|Q8S530|Q8S530 Cytosolic aldehyde dehydrogenase RF2D (Fragme... 310 2e-83
sptr|Q8S529|Q8S529 Cytosolic aldehyde dehydrogenase RF2D. 310 2e-83
sptr|Q9LV57|Q9LV57 Aldehyde dehydrogenase ALDH1a. 280 2e-74
sptr|Q93XI6|Q93XI6 Mitochondrial aldehyde dehydrogenase ALDH2. 250 1e-65
sptr|Q94G64|Q94G64 T-cytoplasm male sterility restorer factor 2. 249 2e-65
sptr|Q43274|Q43274 T CYTOPLASM MALE sterility RESTORER factor 2 ... 249 3e-65
sptr|Q8LST6|Q8LST6 Mitochondrial aldehyde dehydrogenase. 248 9e-65
sptr|Q8LST5|Q8LST5 Mitochondrial aldehyde dehydrogenase. 248 9e-65
sptr|Q8RUR9|Q8RUR9 Mitochondrial aldehyde dehydrogenase RF2B. 247 1e-64
sptr|Q8RVW2|Q8RVW2 Aldehyde dehydrogenase (Fragment). 247 2e-64
sptr|Q8LST4|Q8LST4 Mitochondrial aldehyde dehydrogenase. 246 2e-64
sptr|P93344|P93344 Aldehyde dehydrogenase (NAD+) (EC 1.2.1.3). 245 5e-64
sptr|Q9FRX7|Q9FRX7 Aldehyde dehydrogenase ALDH2b. 245 5e-64
sptr|Q9LLR2|Q9LLR2 Aldehyde dehydrogenase. 245 5e-64
sptr|Q9PWJ3|Q9PWJ3 Aldehyde dehydrogenase class 1 (EC 1.2.1.3). 244 1e-63
sptr|Q84V96|Q84V96 Aldehyde dehydrogenase 1 precursor (EC 1.2.1.3). 243 3e-63
sptr|Q9YGY2|Q9YGY2 Aldehyde dehydrogenase (EC 1.2.1.3). 242 4e-63
sptr|Q7SXU3|Q7SXU3 Aldehyde dehydrogenase 2. 241 7e-63
sptr|Q9W617|Q9W617 Aldehyde dehydrogenase class 1 (EC 1.2.1.3). 240 2e-62
sptr|Q9SU63|Q9SU63 Aldehyde dehydrogenase (NAD+)-like protein (E... 240 2e-62
sptr|Q8QGQ2|Q8QGQ2 Aldehyde dehydrogenase 2. 239 3e-62
sptr|Q94C67|Q94C67 Putative aldehyde dehydrogenase NAD+. 238 6e-62
sptr|Q8S528|Q8S528 Mitochondrial aldehyde dehydrogenase. 238 6e-62
sptr|Q9ZUB6|Q9ZUB6 F5O8.35 protein. 238 6e-62
sptr|Q9LRI6|Q9LRI6 Mitochondrial aldehyde dehydrogenase ALDH2a. 238 7e-62
sptrnew|AAQ97741|AAQ97741 Mitochondrial aldehyde dehydrogenase 2... 237 2e-61
>sptr|Q8S531|Q8S531 Cytosolic aldehyde dehydrogenase RF2C.
Length = 503
Score = 409 bits (1050), Expect = e-113
Identities = 200/200 (100%), Positives = 200/200 (100%)
Frame = +3
Query: 3 VAGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKR 182
VAGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKR
Sbjct: 304 VAGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKR 363
Query: 183 EGATLVTGGKPCGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKAN 362
EGATLVTGGKPCGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKAN
Sbjct: 364 EGATLVTGGKPCGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKAN 423
Query: 363 NTRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDA 542
NTRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDA
Sbjct: 424 NTRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDA 483
Query: 543 LDKYLQTKTVVTPLYNTPWL 602
LDKYLQTKTVVTPLYNTPWL
Sbjct: 484 LDKYLQTKTVVTPLYNTPWL 503
>sptr|Q8S532|Q8S532 Cytosolic aldehyde dehydrogenase RF2C.
Length = 502
Score = 402 bits (1032), Expect = e-111
Identities = 199/200 (99%), Positives = 199/200 (99%)
Frame = +3
Query: 3 VAGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKR 182
VAGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKR
Sbjct: 304 VAGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKR 363
Query: 183 EGATLVTGGKPCGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKAN 362
EGATLVTGGKPCGD KGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKAN
Sbjct: 364 EGATLVTGGKPCGD-KGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKAN 422
Query: 363 NTRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDA 542
NTRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDA
Sbjct: 423 NTRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDA 482
Query: 543 LDKYLQTKTVVTPLYNTPWL 602
LDKYLQTKTVVTPLYNTPWL
Sbjct: 483 LDKYLQTKTVVTPLYNTPWL 502
>sptr|Q9LRE9|Q9LRE9 Cytosolic aldehyde dehydrogenase.
Length = 502
Score = 365 bits (937), Expect = e-100
Identities = 175/200 (87%), Positives = 188/200 (94%)
Frame = +3
Query: 3 VAGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKR 182
VAG+RIYVQEGIYD FVKKA E+A KSVVGDPFNP V QGPQ+DK+QYEK+L+YIDIGKR
Sbjct: 304 VAGSRIYVQEGIYDAFVKKATEMAKKSVVGDPFNPRVHQGPQIDKEQYEKILKYIDIGKR 363
Query: 183 EGATLVTGGKPCGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKAN 362
EGATLVTGGKPCG+N GYYIEPTIFTDVK++M+IAQ+EIFGPVMALMKFKTVEE IQKAN
Sbjct: 364 EGATLVTGGKPCGEN-GYYIEPTIFTDVKEEMSIAQEEIFGPVMALMKFKTVEEAIQKAN 422
Query: 363 NTRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDA 542
+TRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCY FDPD PFGGYKMSGFGKDMGMDA
Sbjct: 423 STRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYLGFDPDVPFGGYKMSGFGKDMGMDA 482
Query: 543 LDKYLQTKTVVTPLYNTPWL 602
L+KYL TK VVTPLYNTPWL
Sbjct: 483 LEKYLHTKAVVTPLYNTPWL 502
>sptr|Q94JC6|Q94JC6 Putative cytosolic aldehyde dehydrogenase.
Length = 507
Score = 315 bits (808), Expect = 4e-85
Identities = 148/199 (74%), Positives = 176/199 (88%)
Frame = +3
Query: 3 VAGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKR 182
VAG+R+YVQEGIYDEFVKKA E A VGDPF+ + + GPQVDK Q+E+VL+YI+IGK
Sbjct: 309 VAGSRVYVQEGIYDEFVKKAVEAAKNWKVGDPFDAATNMGPQVDKVQFERVLKYIEIGKN 368
Query: 183 EGATLVTGGKPCGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKAN 362
EGATL+TGGKP GD KGYYIEPTIF DVK++MTIAQ+EIFGPVM+LMKFKTVEE I+KAN
Sbjct: 369 EGATLLTGGKPTGD-KGYYIEPTIFVDVKEEMTIAQEEIFGPVMSLMKFKTVEEAIEKAN 427
Query: 363 NTRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDA 542
T+YGLAAGIVTKN+++AN VSRS+RAG +W+NCYFAFDPDAPFGGYKMSGFG+D GM A
Sbjct: 428 CTKYGLAAGIVTKNLNIANMVSRSVRAGTVWVNCYFAFDPDAPFGGYKMSGFGRDQGMVA 487
Query: 543 LDKYLQTKTVVTPLYNTPW 599
+DKYLQ KTV+T + ++PW
Sbjct: 488 MDKYLQVKTVITAVPDSPW 506
>sptr|Q8S530|Q8S530 Cytosolic aldehyde dehydrogenase RF2D
(Fragment).
Length = 466
Score = 310 bits (794), Expect = 2e-83
Identities = 147/199 (73%), Positives = 174/199 (87%)
Frame = +3
Query: 3 VAGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKR 182
VAG+R+YVQEGIYDEFVKKA E A VGDPF+ + + GPQVDKDQ+E+VL+YI+ GK
Sbjct: 268 VAGSRVYVQEGIYDEFVKKAVEAARSWKVGDPFDVTSNMGPQVDKDQFERVLKYIEHGKS 327
Query: 183 EGATLVTGGKPCGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKAN 362
EGATL+TGGKP D KGYYIEPTIF DV +DM IAQ+EIFGPVM+LMKFKTV+EVI+KAN
Sbjct: 328 EGATLLTGGKPAAD-KGYYIEPTIFVDVTEDMKIAQEEIFGPVMSLMKFKTVDEVIEKAN 386
Query: 363 NTRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDA 542
TRYGLAAGIVTK++DVAN VSRS+RAG +W+NCYFAFDPDAPFGGYKMSGFG+D G+ A
Sbjct: 387 CTRYGLAAGIVTKSLDVANRVSRSVRAGTVWVNCYFAFDPDAPFGGYKMSGFGRDQGLAA 446
Query: 543 LDKYLQTKTVVTPLYNTPW 599
+DKYLQ K+V+T L ++PW
Sbjct: 447 MDKYLQVKSVITALPDSPW 465
>sptr|Q8S529|Q8S529 Cytosolic aldehyde dehydrogenase RF2D.
Length = 511
Score = 310 bits (794), Expect = 2e-83
Identities = 147/199 (73%), Positives = 174/199 (87%)
Frame = +3
Query: 3 VAGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKR 182
VAG+R+YVQEGIYDEFVKKA E A VGDPF+ + + GPQVDKDQ+E+VL+YI+ GK
Sbjct: 313 VAGSRVYVQEGIYDEFVKKAVEAARSWKVGDPFDVTSNMGPQVDKDQFERVLKYIEHGKS 372
Query: 183 EGATLVTGGKPCGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKAN 362
EGATL+TGGKP D KGYYIEPTIF DV +DM IAQ+EIFGPVM+LMKFKTV+EVI+KAN
Sbjct: 373 EGATLLTGGKPAAD-KGYYIEPTIFVDVTEDMKIAQEEIFGPVMSLMKFKTVDEVIEKAN 431
Query: 363 NTRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDA 542
TRYGLAAGIVTK++DVAN VSRS+RAG +W+NCYFAFDPDAPFGGYKMSGFG+D G+ A
Sbjct: 432 CTRYGLAAGIVTKSLDVANRVSRSVRAGTVWVNCYFAFDPDAPFGGYKMSGFGRDQGLAA 491
Query: 543 LDKYLQTKTVVTPLYNTPW 599
+DKYLQ K+V+T L ++PW
Sbjct: 492 MDKYLQVKSVITALPDSPW 510
>sptr|Q9LV57|Q9LV57 Aldehyde dehydrogenase ALDH1a.
Length = 501
Score = 280 bits (715), Expect = 2e-74
Identities = 134/200 (67%), Positives = 165/200 (82%)
Frame = +3
Query: 3 VAGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKR 182
VA +R++VQEGIYD+ V+K E A VGDPF+ + QGPQVDK Q+EK+L YI+ GK
Sbjct: 303 VASSRVFVQEGIYDKVVEKLVEKAKDWTVGDPFDSTARQGPQVDKRQFEKILSYIEHGKN 362
Query: 183 EGATLVTGGKPCGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKAN 362
EGATL+TGGK GD KGY+I+PTIF DV +DM I QDEIFGPVM+LMKFKTVEE I+ AN
Sbjct: 363 EGATLLTGGKAIGD-KGYFIQPTIFADVTEDMKIYQDEIFGPVMSLMKFKTVEEGIKCAN 421
Query: 363 NTRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDA 542
NT+YGLAAGI++++ID+ NTVSRSI+AG IW+NCYF FD D P+GGYKMSG ++ GMDA
Sbjct: 422 NTKYGLAAGILSQDIDLINTVSRSIKAGIIWVNCYFGFDLDCPYGGYKMSGNCRESGMDA 481
Query: 543 LDKYLQTKTVVTPLYNTPWL 602
LD YLQTK+VV PL+N+PW+
Sbjct: 482 LDNYLQTKSVVMPLHNSPWM 501
>sptr|Q93XI6|Q93XI6 Mitochondrial aldehyde dehydrogenase ALDH2.
Length = 549
Score = 250 bits (639), Expect = 1e-65
Identities = 118/199 (59%), Positives = 151/199 (75%)
Frame = +3
Query: 6 AGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKRE 185
AG+R +V E +YDEFV+K+ A K VVGDPF V QGPQ+D +Q++K+LRYI G
Sbjct: 352 AGSRTFVHERVYDEFVEKSKARALKRVVGDPFRKGVEQGPQIDDEQFKKILRYIKSGVDS 411
Query: 186 GATLVTGGKPCGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKANN 365
GATLVTGG GD KGYYI+PTIF+DV+DDM IAQ+EIFGPV ++ KF + EVI++AN
Sbjct: 412 GATLVTGGDKLGD-KGYYIQPTIFSDVQDDMKIAQEEIFGPVQSIFKFNDLNEVIKRANA 470
Query: 366 TRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDAL 545
++YGLAAG+ T N+D ANT++R++RAG IW+NC+ FD PFGGYKMSG G++ G+D+L
Sbjct: 471 SQYGLAAGVFTNNLDTANTLTRALRAGTIWVNCFDIFDAAIPFGGYKMSGIGREKGIDSL 530
Query: 546 DKYLQTKTVVTPLYNTPWL 602
YLQ K VVT L N WL
Sbjct: 531 KNYLQVKAVVTALKNPAWL 549
>sptr|Q94G64|Q94G64 T-cytoplasm male sterility restorer factor 2.
Length = 549
Score = 249 bits (637), Expect = 2e-65
Identities = 116/199 (58%), Positives = 153/199 (76%)
Frame = +3
Query: 6 AGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKRE 185
AG+R +V E +YDEFV+KA A K VVGDPF V QGPQ+D +Q++K+LRYI G
Sbjct: 352 AGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDEQFKKILRYIRYGVDG 411
Query: 186 GATLVTGGKPCGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKANN 365
GATLVTGG GD KG+YI+PTIF+DV+D M IAQ+EIFGPV +++KFK + EVI++AN
Sbjct: 412 GATLVTGGDRLGD-KGFYIQPTIFSDVQDGMKIAQEEIFGPVQSILKFKDLNEVIKRANA 470
Query: 366 TRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDAL 545
++YGLAAG+ T ++D ANT++R++RAG +W+NC+ FD PFGGYKMSG G++ G+D+L
Sbjct: 471 SQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGYKMSGMGREKGVDSL 530
Query: 546 DKYLQTKTVVTPLYNTPWL 602
YLQ K VVTP+ N WL
Sbjct: 531 KNYLQVKAVVTPIKNAAWL 549
>sptr|Q43274|Q43274 T CYTOPLASM MALE sterility RESTORER factor 2 (EC
1.2.1.3) (Aldehyde dehydrogenase (NAD+)).
Length = 549
Score = 249 bits (636), Expect = 3e-65
Identities = 116/199 (58%), Positives = 152/199 (76%)
Frame = +3
Query: 6 AGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKRE 185
AG+R +V E +YDEFV+KA A K VVGDPF V QGPQ+D +Q+ K+LRYI G
Sbjct: 352 AGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDEQFNKILRYIRYGVDG 411
Query: 186 GATLVTGGKPCGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKANN 365
GATLVTGG GD KG+YI+PTIF+DV+D M IAQ+EIFGPV +++KFK + EVI++AN
Sbjct: 412 GATLVTGGDRLGD-KGFYIQPTIFSDVQDGMKIAQEEIFGPVQSILKFKDLNEVIKRANA 470
Query: 366 TRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDAL 545
++YGLAAG+ T ++D ANT++R++RAG +W+NC+ FD PFGGYKMSG G++ G+D+L
Sbjct: 471 SQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGYKMSGIGREKGVDSL 530
Query: 546 DKYLQTKTVVTPLYNTPWL 602
YLQ K VVTP+ N WL
Sbjct: 531 KNYLQVKAVVTPIKNAAWL 549
>sptr|Q8LST6|Q8LST6 Mitochondrial aldehyde dehydrogenase.
Length = 549
Score = 248 bits (632), Expect = 9e-65
Identities = 117/199 (58%), Positives = 150/199 (75%)
Frame = +3
Query: 6 AGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKRE 185
AG+R +V E +YDEFV+K+ A K VVGDPF V QGPQ+D +Q++K+LRYI G
Sbjct: 352 AGSRTFVHERVYDEFVEKSKARALKRVVGDPFRKGVEQGPQIDDEQFKKILRYIKSGVDS 411
Query: 186 GATLVTGGKPCGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKANN 365
GATLVTGG GD KGYYI+PTIF+DV+D M IAQ+EIFGPV ++ KF + EVI++AN
Sbjct: 412 GATLVTGGDKLGD-KGYYIQPTIFSDVQDGMKIAQEEIFGPVQSIFKFNDLNEVIKRANA 470
Query: 366 TRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDAL 545
++YGLAAG+ T N+D ANT++R++RAG IW+NC+ FD PFGGYKMSG G++ G+D+L
Sbjct: 471 SQYGLAAGVFTNNLDTANTLTRALRAGTIWVNCFDIFDAAIPFGGYKMSGIGREKGIDSL 530
Query: 546 DKYLQTKTVVTPLYNTPWL 602
YLQ K VVT L N WL
Sbjct: 531 KNYLQVKAVVTALKNPAWL 549
>sptr|Q8LST5|Q8LST5 Mitochondrial aldehyde dehydrogenase.
Length = 551
Score = 248 bits (632), Expect = 9e-65
Identities = 115/199 (57%), Positives = 150/199 (75%)
Frame = +3
Query: 6 AGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKRE 185
AG+R +V E +YDEFV+K+ A K VVGDPF V QGPQ+D DQ+ K+LRY+ G
Sbjct: 354 AGSRTFVHERVYDEFVEKSKARALKRVVGDPFRNGVEQGPQIDGDQFNKILRYVQSGVDS 413
Query: 186 GATLVTGGKPCGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKANN 365
GATLVTGG G ++G+YI+PT+F D KDDM IA++EIFGPV ++KF +EEVI++AN
Sbjct: 414 GATLVTGGDRVG-SRGFYIQPTVFADAKDDMKIAREEIFGPVQTILKFSGMEEVIRRANA 472
Query: 366 TRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDAL 545
T YGLAAG+ T+++D ANT+SR++RAG +W+NCY FD PFGGYKMSG G++ G+ AL
Sbjct: 473 THYGLAAGVFTRSLDAANTLSRALRAGTVWVNCYDVFDATIPFGGYKMSGVGREKGVYAL 532
Query: 546 DKYLQTKTVVTPLYNTPWL 602
YLQTK VVTP+ + WL
Sbjct: 533 RNYLQTKAVVTPIKDPAWL 551
>sptr|Q8RUR9|Q8RUR9 Mitochondrial aldehyde dehydrogenase RF2B.
Length = 550
Score = 247 bits (631), Expect = 1e-64
Identities = 115/199 (57%), Positives = 149/199 (74%)
Frame = +3
Query: 6 AGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKRE 185
AG+R +V E +YDEFV+K+ A K VVGDPF V QGPQ+D +Q+ K+LRY+ G
Sbjct: 353 AGSRTFVHERVYDEFVEKSKARALKRVVGDPFRDGVEQGPQIDGEQFNKILRYVQSGVDS 412
Query: 186 GATLVTGGKPCGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKANN 365
GATLV GG GD +G+YI+PT+F D KD+M IA++EIFGPV ++KF VEEVI++AN
Sbjct: 413 GATLVAGGDRVGD-RGFYIQPTVFADAKDEMKIAREEIFGPVQTILKFSGVEEVIRRANA 471
Query: 366 TRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDAL 545
T YGLAAG+ T+++D ANT+SR++RAG +W+NCY FD PFGGYKMSG G++ G+ AL
Sbjct: 472 TPYGLAAGVFTRSLDAANTLSRALRAGTVWVNCYDVFDATIPFGGYKMSGVGREKGIYAL 531
Query: 546 DKYLQTKTVVTPLYNTPWL 602
YLQTK VVTP+ N WL
Sbjct: 532 RNYLQTKAVVTPIKNPAWL 550
>sptr|Q8RVW2|Q8RVW2 Aldehyde dehydrogenase (Fragment).
Length = 230
Score = 247 bits (630), Expect = 2e-64
Identities = 116/194 (59%), Positives = 150/194 (77%)
Frame = +3
Query: 21 YVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKREGATLV 200
+V E IYDEFV+KA A + VVGDPF V QGPQ+D +Q+ K+LRYI +G GA+LV
Sbjct: 38 FVHEKIYDEFVEKAKARAMRRVVGDPFKKGVEQGPQIDDEQFNKILRYIKLGVDGGASLV 97
Query: 201 TGGKPCGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKANNTRYGL 380
TGG+ G +KGYYI+PTIF+DVKDDM IA++EIFGPV +++KF + EVI++AN +RYGL
Sbjct: 98 TGGERIG-SKGYYIQPTIFSDVKDDMVIAKEEIFGPVQSILKFNDLNEVIRRANASRYGL 156
Query: 381 AAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDALDKYLQ 560
AAG+ T NID ANT++R++R G +W+NC+ FD PFGGYKMSG G++ G+D+L YLQ
Sbjct: 157 AAGVFTSNIDKANTLTRALRVGTVWVNCFDVFDAAIPFGGYKMSGQGREKGIDSLKGYLQ 216
Query: 561 TKTVVTPLYNTPWL 602
TK VVTPL N WL
Sbjct: 217 TKAVVTPLKNPAWL 230
>sptr|Q8LST4|Q8LST4 Mitochondrial aldehyde dehydrogenase.
Length = 547
Score = 246 bits (629), Expect = 2e-64
Identities = 116/199 (58%), Positives = 150/199 (75%)
Frame = +3
Query: 6 AGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKRE 185
AG+R +V E +YDEFV+KA A K VVGDPF V QGPQ+D +Q+ K+LRYI G
Sbjct: 350 AGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDEQFNKILRYIRSGVDS 409
Query: 186 GATLVTGGKPCGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKANN 365
GA LVTGG G+ KGYYI+PTIF+DV+D M IAQ+EIFGPV +++KFK + EVI++AN
Sbjct: 410 GANLVTGGDRLGE-KGYYIQPTIFSDVQDGMKIAQEEIFGPVQSILKFKDLNEVIKRANA 468
Query: 366 TRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDAL 545
+ YGLAAG+ T ++D ANT++R++RAG +WINC+ FD PFGGYKMSG G++ G+D+L
Sbjct: 469 SPYGLAAGVFTNSLDTANTLTRALRAGTVWINCFDIFDAAIPFGGYKMSGIGREKGIDSL 528
Query: 546 DKYLQTKTVVTPLYNTPWL 602
YLQ K VVTP+ N WL
Sbjct: 529 KNYLQVKAVVTPIKNAAWL 547
>sptr|P93344|P93344 Aldehyde dehydrogenase (NAD+) (EC 1.2.1.3).
Length = 542
Score = 245 bits (626), Expect = 5e-64
Identities = 113/199 (56%), Positives = 150/199 (75%)
Frame = +3
Query: 6 AGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKRE 185
AG+R +V E +YDEF++KA A K VGDPF QGPQ+D Q++K++ YI G
Sbjct: 345 AGSRTFVHEKVYDEFLEKAKARALKRTVGDPFKSGTEQGPQIDSKQFDKIMNYIRSGIDS 404
Query: 186 GATLVTGGKPCGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKANN 365
GATL TGG+ G+ +GYYI+PT+F++VKDDM IAQDEIFGPV +++KFK V++VI++ANN
Sbjct: 405 GATLETGGERLGE-RGYYIKPTVFSNVKDDMLIAQDEIFGPVQSILKFKDVDDVIRRANN 463
Query: 366 TRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDAL 545
+RYGLAAG+ T+NID ANT++R++R G +W+NC+ FD PFGGYKMSG G++ G +L
Sbjct: 464 SRYGLAAGVFTQNIDTANTLTRALRVGTVWVNCFDTFDATIPFGGYKMSGHGREKGEYSL 523
Query: 546 DKYLQTKTVVTPLYNTPWL 602
YLQ K VVTPL N WL
Sbjct: 524 KNYLQVKAVVTPLKNPAWL 542
>sptr|Q9FRX7|Q9FRX7 Aldehyde dehydrogenase ALDH2b.
Length = 549
Score = 245 bits (626), Expect = 5e-64
Identities = 114/199 (57%), Positives = 149/199 (74%)
Frame = +3
Query: 6 AGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKRE 185
AG+R +V E IYDEFV+KA A K VVGDPF V QGPQ+D +Q+ K+LRYI G
Sbjct: 352 AGSRTFVHERIYDEFVEKAKARALKRVVGDPFKNGVEQGPQIDDEQFNKILRYIKYGVDS 411
Query: 186 GATLVTGGKPCGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKANN 365
GA LVTGG GD KGYYI+PTIF+DV+D+M IAQ+EIFGPV +++KF + EVI++AN
Sbjct: 412 GANLVTGGDRLGD-KGYYIQPTIFSDVQDNMRIAQEEIFGPVQSILKFNDLNEVIKRANA 470
Query: 366 TRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDAL 545
++YGLAAG+ T N++ ANT++R++R G +W+NC+ FD PFGGYK SG G++ G+D+L
Sbjct: 471 SQYGLAAGVFTNNLNTANTLTRALRVGTVWVNCFDVFDAAIPFGGYKQSGIGREKGIDSL 530
Query: 546 DKYLQTKTVVTPLYNTPWL 602
YLQ K VVTP+ N WL
Sbjct: 531 KNYLQVKAVVTPIKNAAWL 549
>sptr|Q9LLR2|Q9LLR2 Aldehyde dehydrogenase.
Length = 549
Score = 245 bits (626), Expect = 5e-64
Identities = 114/199 (57%), Positives = 149/199 (74%)
Frame = +3
Query: 6 AGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKRE 185
AG+R +V E IYDEFV+KA A K VVGDPF V QGPQ+D +Q+ K+LRYI G
Sbjct: 352 AGSRTFVHERIYDEFVEKAKARALKRVVGDPFKNGVEQGPQIDDEQFNKILRYIKYGVDS 411
Query: 186 GATLVTGGKPCGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKANN 365
GA LVTGG GD KGYYI+PTIF+DV+D+M IAQ+EIFGPV +++KF + EVI++AN
Sbjct: 412 GANLVTGGDRLGD-KGYYIQPTIFSDVQDNMRIAQEEIFGPVQSILKFNDLNEVIKRANA 470
Query: 366 TRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDAL 545
++YGLAAG+ T N++ ANT++R++R G +W+NC+ FD PFGGYK SG G++ G+D+L
Sbjct: 471 SQYGLAAGVFTNNLNTANTLTRALRVGTVWVNCFDVFDAAIPFGGYKQSGIGREKGIDSL 530
Query: 546 DKYLQTKTVVTPLYNTPWL 602
YLQ K VVTP+ N WL
Sbjct: 531 KNYLQVKAVVTPIKNAAWL 549
>sptr|Q9PWJ3|Q9PWJ3 Aldehyde dehydrogenase class 1 (EC 1.2.1.3).
Length = 502
Score = 244 bits (622), Expect = 1e-63
Identities = 111/191 (58%), Positives = 153/191 (80%)
Frame = +3
Query: 3 VAGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKR 182
+AG+RI+V+E IYDEFV+K+ E A K V+GDPF P V+QGPQ+DK+QY+K+L I+ GK+
Sbjct: 305 IAGSRIFVEEPIYDEFVRKSVERAKKRVLGDPFAPCVNQGPQIDKEQYDKILELIESGKK 364
Query: 183 EGATLVTGGKPCGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKAN 362
EGA L GG G+ KG+YI PT+F+DVKDDM IA++EIFGPV ++KFKT++EVI++AN
Sbjct: 365 EGAKLQCGGSAWGE-KGFYISPTVFSDVKDDMRIAKEEIFGPVQQILKFKTIDEVIKRAN 423
Query: 363 NTRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDA 542
NT+YGLAAG+ TK++D A +S +++AG +WINCY A P +PFGG+KMSG G++MG
Sbjct: 424 NTKYGLAAGVFTKDMDKAILMSTALQAGTVWINCYSAMSPQSPFGGFKMSGNGREMGEYG 483
Query: 543 LDKYLQTKTVV 575
L +Y + KTV+
Sbjct: 484 LHEYTEVKTVI 494
>sptr|Q84V96|Q84V96 Aldehyde dehydrogenase 1 precursor (EC 1.2.1.3).
Length = 542
Score = 243 bits (619), Expect = 3e-63
Identities = 112/199 (56%), Positives = 150/199 (75%)
Frame = +3
Query: 6 AGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKRE 185
AG+R +V E +YDEF++K+ + A + VVGDPF V QGPQ+D +Q+EK+LRYI G
Sbjct: 345 AGSRTFVHERVYDEFLEKSKKRALRRVVGDPFKKGVEQGPQIDTEQFEKILRYIKSGIES 404
Query: 186 GATLVTGGKPCGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKANN 365
ATL GG G +KG+Y++PT+F++V+DDM IAQDEIFGPV + KFK ++EVI++AN+
Sbjct: 405 NATLECGGDRLG-SKGFYVQPTVFSNVQDDMLIAQDEIFGPVQTIFKFKEIDEVIRRANS 463
Query: 366 TRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDAL 545
TRYGLAAG+ T+N+ ANT+ R++RAG +WINC+ FD PFGGYKMSG G++ G+ +L
Sbjct: 464 TRYGLAAGVFTQNLATANTLMRALRAGTVWINCFDVFDAAIPFGGYKMSGIGREKGIYSL 523
Query: 546 DKYLQTKTVVTPLYNTPWL 602
YLQ K VVTPL N WL
Sbjct: 524 HNYLQVKAVVTPLKNPAWL 542
>sptr|Q9YGY2|Q9YGY2 Aldehyde dehydrogenase (EC 1.2.1.3).
Length = 502
Score = 242 bits (618), Expect = 4e-63
Identities = 111/191 (58%), Positives = 152/191 (79%)
Frame = +3
Query: 3 VAGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKR 182
+AG+RI+V+E IYDEFV+K+ E A K V+GDPF P V+QGPQ+DK+QY+K L I+ GK+
Sbjct: 305 IAGSRIFVEEPIYDEFVRKSVERAKKRVLGDPFAPCVNQGPQIDKEQYDKCLELIESGKK 364
Query: 183 EGATLVTGGKPCGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKAN 362
EGA L GG G+ KG+YI PT+F+DVKDDM IA++EIFGPV ++KFKT++EVI++AN
Sbjct: 365 EGAKLQCGGSAWGE-KGFYISPTVFSDVKDDMRIAKEEIFGPVQQILKFKTIDEVIKRAN 423
Query: 363 NTRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDA 542
NT+YGLAAG+ TK++D A +S +++AG +WINCY A P +PFGG+KMSG G++MG
Sbjct: 424 NTKYGLAAGVFTKDMDKAILMSTALQAGTVWINCYSAMSPQSPFGGFKMSGNGREMGEYG 483
Query: 543 LDKYLQTKTVV 575
L +Y + KTV+
Sbjct: 484 LHEYTEVKTVI 494
>sptr|Q7SXU3|Q7SXU3 Aldehyde dehydrogenase 2.
Length = 516
Score = 241 bits (616), Expect = 7e-63
Identities = 112/189 (59%), Positives = 148/189 (78%)
Frame = +3
Query: 6 AGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKRE 185
AGTR +VQE IYDEFV+++ E A +VGDPF+ + QGPQVD+DQ++KVL YI GKRE
Sbjct: 320 AGTRTFVQESIYDEFVERSVERAKNRIVGDPFDLNTEQGPQVDEDQFKKVLGYISSGKRE 379
Query: 186 GATLVTGGKPCGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKANN 365
GA L+ GG P + +GY+I+PT+F DVKDDMTIA++EIFGPVM ++KFK++EEVI++AN+
Sbjct: 380 GAKLMCGGAPAAE-RGYFIQPTVFGDVKDDMTIAREEIFGPVMQILKFKSLEEVIERAND 438
Query: 366 TRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDAL 545
++YGLAA + T+NID AN +S +RAG +WINCY F APFGGYK SG G+++G L
Sbjct: 439 SKYGLAAAVFTQNIDKANYISHGLRAGTVWINCYNVFGVQAPFGGYKASGIGRELGEYGL 498
Query: 546 DKYLQTKTV 572
D Y + KTV
Sbjct: 499 DIYTEVKTV 507
>sptr|Q9W617|Q9W617 Aldehyde dehydrogenase class 1 (EC 1.2.1.3).
Length = 414
Score = 240 bits (613), Expect = 2e-62
Identities = 110/191 (57%), Positives = 149/191 (78%)
Frame = +3
Query: 3 VAGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKR 182
+AG+RI+V+E IYDEFV+K+ E A K V+GDP P V QGPQ+DK+QY K+L I+ GK+
Sbjct: 217 IAGSRIFVEEPIYDEFVRKSVERAKKRVLGDPLTPGVCQGPQIDKEQYNKILELIESGKK 276
Query: 183 EGATLVTGGKPCGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKAN 362
EGA L GG G+ KG+YI PT+F+DVKDDM IA++EIFGPV ++KFKT++EVI++AN
Sbjct: 277 EGAKLECGGSAWGE-KGFYISPTVFSDVKDDMRIAKEEIFGPVQQILKFKTIDEVIKRAN 335
Query: 363 NTRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDA 542
NT YGLAAG+ TK++D A +S +++AG +WINCY A P +PFGG+KMSG G++MG
Sbjct: 336 NTNYGLAAGVFTKDMDKAILMSTALQAGTVWINCYSAMSPQSPFGGFKMSGNGREMGEYG 395
Query: 543 LDKYLQTKTVV 575
L +Y + KTV+
Sbjct: 396 LHEYTEVKTVI 406
>sptr|Q9SU63|Q9SU63 Aldehyde dehydrogenase (NAD+)-like protein (EC
1.2.1.3) (Putative aldehyde dehydrogenase (NAD+)
protein) (AT3g48000/T17F15_130) (Aldehyde dehydrogenase
ALDH2a) (NAD+).
Length = 538
Score = 240 bits (612), Expect = 2e-62
Identities = 112/199 (56%), Positives = 148/199 (74%)
Frame = +3
Query: 6 AGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKRE 185
AG+R +V E +YDEFV+K+ A K VVGDPF + QGPQ+D Q+EKV++YI G
Sbjct: 341 AGSRTFVHEKVYDEFVEKSKARALKRVVGDPFRKGIEQGPQIDLKQFEKVMKYIKSGIES 400
Query: 186 GATLVTGGKPCGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKANN 365
ATL GG GD KGY+I+PT+F++VKDDM IAQDEIFGPV +++KF V+EVI++AN
Sbjct: 401 NATLECGGDQIGD-KGYFIQPTVFSNVKDDMLIAQDEIFGPVQSILKFSDVDEVIKRANE 459
Query: 366 TRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDAL 545
T+YGLAAG+ TKN+D AN VSR+++AG +W+NC+ FD PFGGYKMSG G++ G+ +L
Sbjct: 460 TKYGLAAGVFTKNLDTANRVSRALKAGTVWVNCFDVFDAAIPFGGYKMSGNGREKGIYSL 519
Query: 546 DKYLQTKTVVTPLYNTPWL 602
+ YLQ K VVT L W+
Sbjct: 520 NNYLQIKAVVTALNKPAWI 538
>sptr|Q8QGQ2|Q8QGQ2 Aldehyde dehydrogenase 2.
Length = 516
Score = 239 bits (610), Expect = 3e-62
Identities = 111/189 (58%), Positives = 147/189 (77%)
Frame = +3
Query: 6 AGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKRE 185
AGTR +VQE IYDEFV+++ E A +VGDPF+ + QGPQVD+DQ++KVL YI GKRE
Sbjct: 320 AGTRTFVQESIYDEFVERSVERAKNRIVGDPFDLNTEQGPQVDEDQFKKVLGYISSGKRE 379
Query: 186 GATLVTGGKPCGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKANN 365
GA L+ GG P + +GY+I+PT+F DVKDDM IA++EIFGPVM ++KFK++EEVI++AN+
Sbjct: 380 GAKLMCGGAPAAE-RGYFIQPTVFGDVKDDMKIAREEIFGPVMQILKFKSLEEVIERAND 438
Query: 366 TRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDAL 545
++YGLAA + T+NID AN +S +RAG +WINCY F APFGGYK SG G+++G L
Sbjct: 439 SKYGLAAAVFTQNIDKANYISHGLRAGTVWINCYNVFGVQAPFGGYKASGIGRELGEYGL 498
Query: 546 DKYLQTKTV 572
D Y + KTV
Sbjct: 499 DIYTEVKTV 507
>sptr|Q94C67|Q94C67 Putative aldehyde dehydrogenase NAD+.
Length = 534
Score = 238 bits (608), Expect = 6e-62
Identities = 112/199 (56%), Positives = 145/199 (72%)
Frame = +3
Query: 6 AGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKRE 185
AG+R +V E +YDEFV+KA A K VGDPF + QGPQVD +Q+ K+L+YI G
Sbjct: 337 AGSRTFVHERVYDEFVEKAKARALKRNVGDPFKSGIEQGPQVDSEQFNKILKYIKHGVEA 396
Query: 186 GATLVTGGKPCGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKANN 365
GATL GG G +KGYYI+PT+F+DVKDDM IA DEIFGPV ++KFK ++EVI +ANN
Sbjct: 397 GATLQAGGDRLG-SKGYYIQPTVFSDVKDDMLIATDEIFGPVQTILKFKDLDEVIARANN 455
Query: 366 TRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDAL 545
+RYGLAAG+ T+N+D A+ + R++R G +WINC+ D PFGGYKMSG G++ G+ +L
Sbjct: 456 SRYGLAAGVFTQNLDTAHRLMRALRVGTVWINCFDVLDASIPFGGYKMSGIGREKGIYSL 515
Query: 546 DKYLQTKTVVTPLYNTPWL 602
+ YLQ K VVT L N WL
Sbjct: 516 NNYLQVKAVVTSLKNPAWL 534
>sptr|Q8S528|Q8S528 Mitochondrial aldehyde dehydrogenase.
Length = 534
Score = 238 bits (608), Expect = 6e-62
Identities = 112/199 (56%), Positives = 145/199 (72%)
Frame = +3
Query: 6 AGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKRE 185
AG+R +V E +YDEFV+KA A K VGDPF + QGPQVD +Q+ K+L+YI G
Sbjct: 337 AGSRTFVHERVYDEFVEKAKARALKRNVGDPFKSGIEQGPQVDSEQFNKILKYIKHGVEA 396
Query: 186 GATLVTGGKPCGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKANN 365
GATL GG G +KGYYI+PT+F+DVKDDM IA DEIFGPV ++KFK ++EVI +ANN
Sbjct: 397 GATLQAGGDRLG-SKGYYIQPTVFSDVKDDMLIATDEIFGPVQTILKFKDLDEVIARANN 455
Query: 366 TRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDAL 545
+RYGLAAG+ T+N+D A+ + R++R G +WINC+ D PFGGYKMSG G++ G+ +L
Sbjct: 456 SRYGLAAGVFTQNLDTAHRLMRALRVGTVWINCFDVLDASIPFGGYKMSGIGREKGIYSL 515
Query: 546 DKYLQTKTVVTPLYNTPWL 602
+ YLQ K VVT L N WL
Sbjct: 516 NNYLQVKAVVTSLKNPAWL 534
>sptr|Q9ZUB6|Q9ZUB6 F5O8.35 protein.
Length = 519
Score = 238 bits (608), Expect = 6e-62
Identities = 112/199 (56%), Positives = 145/199 (72%)
Frame = +3
Query: 6 AGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKRE 185
AG+R +V E +YDEFV+KA A K VGDPF + QGPQVD +Q+ K+L+YI G
Sbjct: 322 AGSRTFVHERVYDEFVEKAKARALKRNVGDPFKSGIEQGPQVDSEQFNKILKYIKHGVEA 381
Query: 186 GATLVTGGKPCGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKANN 365
GATL GG G +KGYYI+PT+F+DVKDDM IA DEIFGPV ++KFK ++EVI +ANN
Sbjct: 382 GATLQAGGDRLG-SKGYYIQPTVFSDVKDDMLIATDEIFGPVQTILKFKDLDEVIARANN 440
Query: 366 TRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDAL 545
+RYGLAAG+ T+N+D A+ + R++R G +WINC+ D PFGGYKMSG G++ G+ +L
Sbjct: 441 SRYGLAAGVFTQNLDTAHRLMRALRVGTVWINCFDVLDASIPFGGYKMSGIGREKGIYSL 500
Query: 546 DKYLQTKTVVTPLYNTPWL 602
+ YLQ K VVT L N WL
Sbjct: 501 NNYLQVKAVVTSLKNPAWL 519
>sptr|Q9LRI6|Q9LRI6 Mitochondrial aldehyde dehydrogenase ALDH2a.
Length = 553
Score = 238 bits (607), Expect = 7e-62
Identities = 109/199 (54%), Positives = 150/199 (75%)
Frame = +3
Query: 6 AGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKRE 185
AG+R +V E +YDEFV+KA A + VVGDPF V QGPQ+D +Q++K+L+Y+ G
Sbjct: 356 AGSRTFVHERVYDEFVEKARARALQRVVGDPFRTGVEQGPQIDGEQFKKILQYVKSGVDS 415
Query: 186 GATLVTGGKPCGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKANN 365
GATLV GG G ++G+YI+PT+F DV+D+M IAQ+EIFGPV +++KF TVEEV+++AN
Sbjct: 416 GATLVAGGDRAG-SRGFYIQPTVFADVEDEMKIAQEEIFGPVQSILKFSTVEEVVRRANA 474
Query: 366 TRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDAL 545
T YGLAAG+ T+ +D ANT++R++R G +W+N Y FD PFGGYKMSG G++ G+ +L
Sbjct: 475 TPYGLAAGVFTQRLDAANTLARALRVGTVWVNTYDVFDAAVPFGGYKMSGVGREKGVYSL 534
Query: 546 DKYLQTKTVVTPLYNTPWL 602
YLQTK VVTP+ + WL
Sbjct: 535 RNYLQTKAVVTPIKDAAWL 553
>sptrnew|AAQ97741|AAQ97741 Mitochondrial aldehyde dehydrogenase 2
family.
Length = 516
Score = 237 bits (604), Expect = 2e-61
Identities = 109/189 (57%), Positives = 147/189 (77%)
Frame = +3
Query: 6 AGTRIYVQEGIYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKRE 185
AGTR +VQE IYDEFV+++ E A +VGDPF+ + QGPQV++DQ++KVL YI GKRE
Sbjct: 320 AGTRTFVQESIYDEFVERSVERAKNRIVGDPFDLNTEQGPQVNEDQFKKVLGYISSGKRE 379
Query: 186 GATLVTGGKPCGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKANN 365
GA L+ GG P + +GY+I+PT+F DVKDDMTIA++EIFGPVM ++KFK++EEVI++AN+
Sbjct: 380 GAKLMCGGAPAAE-RGYFIQPTVFGDVKDDMTIAREEIFGPVMQILKFKSLEEVIERAND 438
Query: 366 TRYGLAAGIVTKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDAL 545
++YGLA + T++ID AN +S +RAG +WINCY F APFGGYK SG G++MG L
Sbjct: 439 SKYGLAGAVFTQDIDKANYISHGLRAGTVWINCYNVFGVQAPFGGYKASGIGREMGEYGL 498
Query: 546 DKYLQTKTV 572
+ Y + KTV
Sbjct: 499 ENYTEVKTV 507
Database: /db/uniprot/tmp/swall
Posted date: Mar 5, 2004 7:45 PM
Number of letters in database: 442,889,342
Number of sequences in database: 1,395,590
Lambda K H
0.315 0.125 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 574,631,503
Number of Sequences: 1395590
Number of extensions: 12913124
Number of successful extensions: 44377
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 40961
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 42996
length of database: 442,889,342
effective HSP length: 121
effective length of database: 274,022,952
effective search space used: 39733328040
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)

