BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= 2161200.2.2
(1078 letters)
Database: /db/uniprot/tmp/swall
1,395,590 sequences; 442,889,342 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
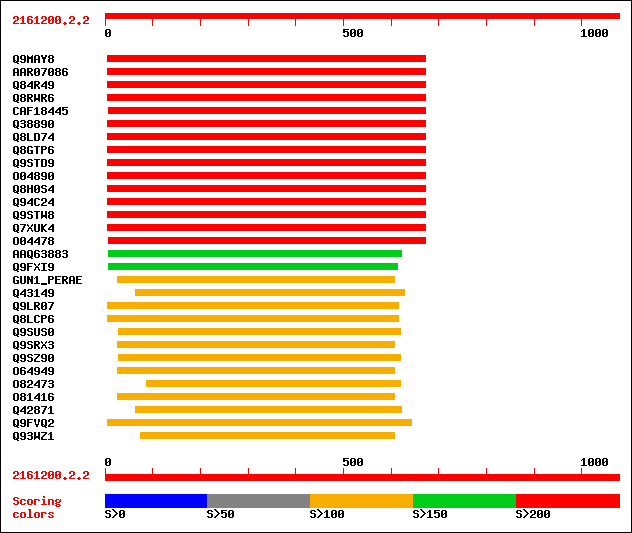
sptr|Q9MAY8|Q9MAY8 Endo-1,4-beta-glucanase Cel1. 427 e-118
sptrnew|AAR07086|AAR07086 Putative endo-1,4-beta-glucanase. 424 e-117
sptr|Q84R49|Q84R49 Putative endo-1,4-beta-glucanase. 424 e-117
sptr|Q8RWR6|Q8RWR6 Endo-1,4-beta-glucanase. 420 e-116
sptrnew|CAF18445|CAF18445 Endo-1,4-beta-D-glucanase KORRIGAN (EC... 377 e-103
sptr|Q38890|Q38890 Cellulase precursor (Cellulase homolog OR16PE... 377 e-103
sptr|Q8LD74|Q8LD74 Cellulase homolog OR16pep. 377 e-103
sptr|Q8GTP6|Q8GTP6 Endo-1,4-beta-D-glucanase (EC 3.2.1.4). 376 e-103
sptr|Q9STD9|Q9STD9 Cellulase (EC 3.2.1.4). 375 e-103
sptr|O04890|O04890 Endo-1,4-beta-glucanase (EC 3.2.1.4). 374 e-102
sptr|Q8H0S4|Q8H0S4 At5g49720/K2I5_8. 373 e-102
sptr|Q94C24|Q94C24 AT5g49720/K2I5_8. 373 e-102
sptr|Q9STW8|Q9STW8 Endo-1, 4-beta-glucanase like protein. 343 3e-93
sptr|Q7XUK4|Q7XUK4 OSJNBa0067K08.14 protein. 313 4e-84
sptr|O04478|O04478 F5I14.14. 276 5e-73
sptrnew|AAQ63883|AAQ63883 Cellulase. 198 1e-49
sptr|Q9FXI9|Q9FXI9 F6F9.1 protein (At1g19940/F6F9_1). 152 1e-35
sw|P05522|GUN1_PERAE Endoglucanase 1 precursor (EC 3.2.1.4) (End... 149 7e-35
sptr|Q43149|Q43149 Cellulase (EC 3.2.1.4) (Fragment). 147 4e-34
sptr|Q9LR07|Q9LR07 F10A5.13 (Hypothetical protein) (Putative end... 145 8e-34
sptr|Q8LCP6|Q8LCP6 Endo-beta-1,4-glucanase, putative. 145 8e-34
sptr|Q9SUS0|Q9SUS0 Putative cellulase. 145 1e-33
sptr|Q9SRX3|Q9SRX3 Endo-1,4-beta glucanase. 145 1e-33
sptr|Q9SZ90|Q9SZ90 Cellulase-like protein. 145 1e-33
sptr|O64949|O64949 Endo-1,4-beta glucanase. 144 2e-33
sptr|O82473|O82473 Endo-1,4-beta-glucanase (EC 3.2.1.4). 144 3e-33
sptr|O81416|O81416 T2H3.5 protein (Putative endo-1, 4-beta gluca... 142 1e-32
sptr|Q42871|Q42871 Endo-1,4-beta-glucanase precursor (EC 3.2.1.4). 142 1e-32
sptr|Q9FVQ2|Q9FVQ2 Endo-beta-1,4-glucanase, putative. 141 2e-32
sptr|Q93WZ1|Q93WZ1 Endo-beta-1,4-glucanase precursor (EC 3.2.1.4). 141 2e-32
>sptr|Q9MAY8|Q9MAY8 Endo-1,4-beta-glucanase Cel1.
Length = 621
Score = 427 bits (1098), Expect = e-118
Identities = 198/223 (88%), Positives = 214/223 (95%)
Frame = +2
Query: 5 EVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPLFNSFNFTKGGLIQLNHGRPQPL 184
+VLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLP+FNSFNFTKGGLIQLNHG PQPL
Sbjct: 383 QVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPVFNSFNFTKGGLIQLNHGGPQPL 442
Query: 185 QYAVNAAFLASLYSDYLEAADTPGWYCGPNFYTTEVLRKFARSQLDYILGKNPLKMSYVV 364
QY VNAAFLASLY+DYL+ ADTPGWYCGPNFYTT+VLRKFA+SQLDYILGKNP KMSYVV
Sbjct: 443 QYVVNAAFLASLYADYLDTADTPGWYCGPNFYTTDVLRKFAKSQLDYILGKNPQKMSYVV 502
Query: 365 GFGNKYPKRPHHRGASIPHNGVKYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDV 544
GFG KYPKR HHRGASIPHNGVKYGCKGG+KWR++KKANPNILVGAMVAGPD+ DG+KD+
Sbjct: 503 GFGKKYPKRVHHRGASIPHNGVKYGCKGGFKWRESKKANPNILVGAMVAGPDKHDGFKDI 562
Query: 545 RTNYNYTEPTLAANAGLVAALISISDIKTGRFGIDKNTIFSAI 673
RTNYNYTEPTLAANAGLVAALIS++DI TGR+ IDKNTIFSA+
Sbjct: 563 RTNYNYTEPTLAANAGLVAALISLADIDTGRYSIDKNTIFSAV 605
>sptrnew|AAR07086|AAR07086 Putative endo-1,4-beta-glucanase.
Length = 620
Score = 424 bits (1089), Expect = e-117
Identities = 198/223 (88%), Positives = 214/223 (95%)
Frame = +2
Query: 5 EVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPLFNSFNFTKGGLIQLNHGRPQPL 184
+VLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLP++NSFNFTKGG+IQLNHGRPQPL
Sbjct: 383 QVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPMYNSFNFTKGGMIQLNHGRPQPL 442
Query: 185 QYAVNAAFLASLYSDYLEAADTPGWYCGPNFYTTEVLRKFARSQLDYILGKNPLKMSYVV 364
QY VNAAFLASLYSDYL+AADTPGWYCGP FYTTEVLRKFARSQLDY+LGKNPLKMSYVV
Sbjct: 443 QYVVNAAFLASLYSDYLDAADTPGWYCGPTFYTTEVLRKFARSQLDYVLGKNPLKMSYVV 502
Query: 365 GFGNKYPKRPHHRGASIPHNGVKYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDV 544
GFGNKYPKR HHRGASIPHNGVKYGCKGG+KWR+TKK NPNIL+GA+VAGPDR DG+KDV
Sbjct: 503 GFGNKYPKRAHHRGASIPHNGVKYGCKGGFKWRETKKPNPNILIGALVAGPDRHDGFKDV 562
Query: 545 RTNYNYTEPTLAANAGLVAALISISDIKTGRFGIDKNTIFSAI 673
RTNYNYTEPTLAANAGLVAALIS+++I + GIDKNTIFSA+
Sbjct: 563 RTNYNYTEPTLAANAGLVAALISLTNIHV-KSGIDKNTIFSAV 604
>sptr|Q84R49|Q84R49 Putative endo-1,4-beta-glucanase.
Length = 620
Score = 424 bits (1089), Expect = e-117
Identities = 198/223 (88%), Positives = 214/223 (95%)
Frame = +2
Query: 5 EVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPLFNSFNFTKGGLIQLNHGRPQPL 184
+VLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLP++NSFNFTKGG+IQLNHGRPQPL
Sbjct: 383 QVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPMYNSFNFTKGGMIQLNHGRPQPL 442
Query: 185 QYAVNAAFLASLYSDYLEAADTPGWYCGPNFYTTEVLRKFARSQLDYILGKNPLKMSYVV 364
QY VNAAFLASLYSDYL+AADTPGWYCGP FYTTEVLRKFARSQLDY+LGKNPLKMSYVV
Sbjct: 443 QYVVNAAFLASLYSDYLDAADTPGWYCGPTFYTTEVLRKFARSQLDYVLGKNPLKMSYVV 502
Query: 365 GFGNKYPKRPHHRGASIPHNGVKYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDV 544
GFGNKYPKR HHRGASIPHNGVKYGCKGG+KWR+TKK NPNIL+GA+VAGPDR DG+KDV
Sbjct: 503 GFGNKYPKRAHHRGASIPHNGVKYGCKGGFKWRETKKPNPNILIGALVAGPDRHDGFKDV 562
Query: 545 RTNYNYTEPTLAANAGLVAALISISDIKTGRFGIDKNTIFSAI 673
RTNYNYTEPTLAANAGLVAALIS+++I + GIDKNTIFSA+
Sbjct: 563 RTNYNYTEPTLAANAGLVAALISLTNIHV-KSGIDKNTIFSAV 604
>sptr|Q8RWR6|Q8RWR6 Endo-1,4-beta-glucanase.
Length = 620
Score = 420 bits (1079), Expect = e-116
Identities = 196/223 (87%), Positives = 211/223 (94%)
Frame = +2
Query: 5 EVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPLFNSFNFTKGGLIQLNHGRPQPL 184
+VLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLP+FNSFNFTKGGLIQLNHG PQPL
Sbjct: 383 QVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPVFNSFNFTKGGLIQLNHGGPQPL 442
Query: 185 QYAVNAAFLASLYSDYLEAADTPGWYCGPNFYTTEVLRKFARSQLDYILGKNPLKMSYVV 364
QY VNAAFLASLY+DYL+ ADTP WYC PNFYTT+VLRKFA+SQLDYILGKNP KMSYVV
Sbjct: 443 QYVVNAAFLASLYADYLDTADTPRWYCRPNFYTTDVLRKFAKSQLDYILGKNPQKMSYVV 502
Query: 365 GFGNKYPKRPHHRGASIPHNGVKYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDV 544
GFG KYPKR HHRGASIPHNGVKYGCKGG+KWR+ KKANPNILVGAMVAGPD+ DG+KD+
Sbjct: 503 GFGKKYPKRVHHRGASIPHNGVKYGCKGGFKWREFKKANPNILVGAMVAGPDKHDGFKDI 562
Query: 545 RTNYNYTEPTLAANAGLVAALISISDIKTGRFGIDKNTIFSAI 673
RTNYNYTEPTLAANAGLVAALIS++DI TGR+ IDKNTIFSA+
Sbjct: 563 RTNYNYTEPTLAANAGLVAALISLADIDTGRYSIDKNTIFSAV 605
>sptrnew|CAF18445|CAF18445 Endo-1,4-beta-D-glucanase KORRIGAN (EC
3.2.1.4) (Fragment).
Length = 229
Score = 377 bits (968), Expect = e-103
Identities = 178/222 (80%), Positives = 197/222 (88%)
Frame = +2
Query: 8 VLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPLFNSFNFTKGGLIQLNHGRPQPLQ 187
VLLSRLRLFLSPGYPYEEILRTFHNQT +MCSYLP+F SFN TKGGLIQLNHGRPQPLQ
Sbjct: 1 VLLSRLRLFLSPGYPYEEILRTFHNQTSIIMCSYLPVFTSFNRTKGGLIQLNHGRPQPLQ 60
Query: 188 YAVNAAFLASLYSDYLEAADTPGWYCGPNFYTTEVLRKFARSQLDYILGKNPLKMSYVVG 367
Y VNAAFLA+LYSDYL+AADTPGWYCGPNFY+T+ LR+FAR+Q+DYILGKNP KMSY+VG
Sbjct: 61 YVVNAAFLATLYSDYLDAADTPGWYCGPNFYSTDKLREFARTQIDYILGKNPRKMSYIVG 120
Query: 368 FGNKYPKRPHHRGASIPHNGVKYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDVR 547
FGN YPK HHRGASIP VKY CKGG+KWRDT K NPNILVGAMVAGPDR DG+ DVR
Sbjct: 121 FGNHYPKHVHHRGASIPKTKVKYNCKGGWKWRDTSKPNPNILVGAMVAGPDRHDGFHDVR 180
Query: 548 TNYNYTEPTLAANAGLVAALISISDIKTGRFGIDKNTIFSAI 673
+NYNYTEPTLA NAGLVAAL+++S K+ IDKNT+FSA+
Sbjct: 181 SNYNYTEPTLAGNAGLVAALVALSGDKS--IPIDKNTLFSAV 220
>sptr|Q38890|Q38890 Cellulase precursor (Cellulase homolog OR16PEP
precursor).
Length = 621
Score = 377 bits (967), Expect = e-103
Identities = 176/223 (78%), Positives = 196/223 (87%)
Frame = +2
Query: 5 EVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPLFNSFNFTKGGLIQLNHGRPQPL 184
++LLSRLRLFLSPGYPYEEILRTFHNQT VMCSYLP+FN FN T GGLI+LNHG PQPL
Sbjct: 383 QLLLSRLRLFLSPGYPYEEILRTFHNQTSIVMCSYLPIFNKFNRTNGGLIELNHGAPQPL 442
Query: 185 QYAVNAAFLASLYSDYLEAADTPGWYCGPNFYTTEVLRKFARSQLDYILGKNPLKMSYVV 364
QY+VNAAFLA+LYSDYL+AADTPGWYCGPNFY+T VLR FARSQ+DYILGKNP KMSYVV
Sbjct: 443 QYSVNAAFLATLYSDYLDAADTPGWYCGPNFYSTSVLRDFARSQIDYILGKNPRKMSYVV 502
Query: 365 GFGNKYPKRPHHRGASIPHNGVKYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDV 544
GFG KYP+ HHRGASIP N VKY CKGG+KWRD+KK NPN + GAMVAGPD+RDGY+DV
Sbjct: 503 GFGTKYPRHVHHRGASIPKNKVKYNCKGGWKWRDSKKPNPNTIEGAMVAGPDKRDGYRDV 562
Query: 545 RTNYNYTEPTLAANAGLVAALISISDIKTGRFGIDKNTIFSAI 673
R NYNYTEPTLA NAGLVAAL+++S + IDKNTIFSA+
Sbjct: 563 RMNYNYTEPTLAGNAGLVAALVALSGEEEATGKIDKNTIFSAV 605
>sptr|Q8LD74|Q8LD74 Cellulase homolog OR16pep.
Length = 621
Score = 377 bits (967), Expect = e-103
Identities = 176/223 (78%), Positives = 196/223 (87%)
Frame = +2
Query: 5 EVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPLFNSFNFTKGGLIQLNHGRPQPL 184
++LLSRLRLFLSPGYPYEEILRTFHNQT VMCSYLP+FN FN T GGLI+LNHG PQPL
Sbjct: 383 QLLLSRLRLFLSPGYPYEEILRTFHNQTSIVMCSYLPIFNKFNRTNGGLIELNHGAPQPL 442
Query: 185 QYAVNAAFLASLYSDYLEAADTPGWYCGPNFYTTEVLRKFARSQLDYILGKNPLKMSYVV 364
QY+VNAAFLA+LYSDYL+AADTPGWYCGPNFY+T VLR FARSQ+DYILGKNP KMSYVV
Sbjct: 443 QYSVNAAFLATLYSDYLDAADTPGWYCGPNFYSTSVLRDFARSQIDYILGKNPRKMSYVV 502
Query: 365 GFGNKYPKRPHHRGASIPHNGVKYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDV 544
GFG KYP+ HHRGASIP N VKY CKGG+KWRD+KK NPN + GAMVAGPD+RDGY+DV
Sbjct: 503 GFGTKYPRHVHHRGASIPKNKVKYNCKGGWKWRDSKKPNPNTIEGAMVAGPDKRDGYRDV 562
Query: 545 RTNYNYTEPTLAANAGLVAALISISDIKTGRFGIDKNTIFSAI 673
R NYNYTEPTLA NAGLVAAL+++S + IDKNTIFSA+
Sbjct: 563 RMNYNYTEPTLAGNAGLVAALVALSGEEEATGKIDKNTIFSAV 605
>sptr|Q8GTP6|Q8GTP6 Endo-1,4-beta-D-glucanase (EC 3.2.1.4).
Length = 621
Score = 376 bits (965), Expect = e-103
Identities = 175/223 (78%), Positives = 200/223 (89%)
Frame = +2
Query: 5 EVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPLFNSFNFTKGGLIQLNHGRPQPL 184
+VLLSRL LFLSPGYPYEEILRTFHNQT +MCSYLP+F +FN TKGGLIQLNHGRPQPL
Sbjct: 385 QVLLSRLXLFLSPGYPYEEILRTFHNQTSIIMCSYLPVFTTFNRTKGGLIQLNHGRPQPL 444
Query: 185 QYAVNAAFLASLYSDYLEAADTPGWYCGPNFYTTEVLRKFARSQLDYILGKNPLKMSYVV 364
QY VNAAFLA+LYS+YL+AADTPGWYCGPNFY+T+VLR+FA++Q+DYILGKNP KMSYVV
Sbjct: 445 QYVVNAAFLATLYSEYLDAADTPGWYCGPNFYSTDVLREFAKTQIDYILGKNPRKMSYVV 504
Query: 365 GFGNKYPKRPHHRGASIPHNGVKYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDV 544
GFGN YPK HHRGASIP N +KY CKGG+KWRDT KANPN + GAMVAGPD+ DG++DV
Sbjct: 505 GFGNHYPKHVHHRGASIPKNKIKYSCKGGWKWRDTPKANPNTIDGAMVAGPDKHDGFRDV 564
Query: 545 RTNYNYTEPTLAANAGLVAALISISDIKTGRFGIDKNTIFSAI 673
R+NYNYTEPTLA NAGLVAAL+++S K+ GIDKNTIFSA+
Sbjct: 565 RSNYNYTEPTLAGNAGLVAALVALSGEKS--VGIDKNTIFSAV 605
>sptr|Q9STD9|Q9STD9 Cellulase (EC 3.2.1.4).
Length = 621
Score = 375 bits (962), Expect = e-103
Identities = 174/223 (78%), Positives = 197/223 (88%)
Frame = +2
Query: 5 EVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPLFNSFNFTKGGLIQLNHGRPQPL 184
++LLSRLRLFLSPGYPYEEI+RTFHNQT VMCSYLP FN FN T+GGLI+LNHG PQPL
Sbjct: 383 QLLLSRLRLFLSPGYPYEEIVRTFHNQTSIVMCSYLPYFNKFNRTRGGLIELNHGDPQPL 442
Query: 185 QYAVNAAFLASLYSDYLEAADTPGWYCGPNFYTTEVLRKFARSQLDYILGKNPLKMSYVV 364
QYA NAAFLA+LYSDYL+AADTPGWYCGPNFY+T VLR+FAR+Q+DYILGKNP KMSY+V
Sbjct: 443 QYAANAAFLATLYSDYLDAADTPGWYCGPNFYSTNVLREFARTQIDYILGKNPRKMSYLV 502
Query: 365 GFGNKYPKRPHHRGASIPHNGVKYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDV 544
GFG KYPK HHRGASIP N VKY CKGG+KWRD+KK NPN + GAMVAGPD+RDG++DV
Sbjct: 503 GFGTKYPKHVHHRGASIPKNKVKYNCKGGWKWRDSKKPNPNTIEGAMVAGPDKRDGFRDV 562
Query: 545 RTNYNYTEPTLAANAGLVAALISISDIKTGRFGIDKNTIFSAI 673
RTNYNYTEPTLA NAGLVAAL+++S + IDKNTIFSA+
Sbjct: 563 RTNYNYTEPTLAGNAGLVAALVALSGEEEASGTIDKNTIFSAV 605
>sptr|O04890|O04890 Endo-1,4-beta-glucanase (EC 3.2.1.4).
Length = 617
Score = 374 bits (961), Expect = e-102
Identities = 175/223 (78%), Positives = 199/223 (89%)
Frame = +2
Query: 5 EVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPLFNSFNFTKGGLIQLNHGRPQPL 184
+VLLSR+RLFLSPGYPYEEILRTFHNQT +MCSYLP+F SFN TKGGLIQLNHGRPQPL
Sbjct: 381 QVLLSRMRLFLSPGYPYEEILRTFHNQTSIIMCSYLPIFTSFNRTKGGLIQLNHGRPQPL 440
Query: 185 QYAVNAAFLASLYSDYLEAADTPGWYCGPNFYTTEVLRKFARSQLDYILGKNPLKMSYVV 364
QY VNAAFLA+L+SDYL AADTPGWYCGPNFY+T+VLRKFA +Q+DYILGKNP KMSYVV
Sbjct: 441 QYVVNAAFLATLFSDYLAAADTPGWYCGPNFYSTDVLRKFAETQIDYILGKNPRKMSYVV 500
Query: 365 GFGNKYPKRPHHRGASIPHNGVKYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDV 544
GFGN YPK HHRGASIP N VKY CKGG+K+RD+ KANPN +VGAMVAGPD+ DG++DV
Sbjct: 501 GFGNHYPKHVHHRGASIPKNKVKYNCKGGWKYRDSSKANPNTIVGAMVAGPDKHDGFRDV 560
Query: 545 RTNYNYTEPTLAANAGLVAALISISDIKTGRFGIDKNTIFSAI 673
R+NYNYTEPTLA NAGLVAAL+++S + GIDKNT+FSA+
Sbjct: 561 RSNYNYTEPTLAGNAGLVAALVALSGDRD--VGIDKNTLFSAV 601
>sptr|Q8H0S4|Q8H0S4 At5g49720/K2I5_8.
Length = 621
Score = 373 bits (958), Expect = e-102
Identities = 175/223 (78%), Positives = 195/223 (87%)
Frame = +2
Query: 5 EVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPLFNSFNFTKGGLIQLNHGRPQPL 184
++LLSRLRLFLSPG PYEEILRTFHNQT VMCSYLP+FN FN T GGLI+LNHG PQPL
Sbjct: 383 QLLLSRLRLFLSPGCPYEEILRTFHNQTSIVMCSYLPIFNKFNRTNGGLIELNHGAPQPL 442
Query: 185 QYAVNAAFLASLYSDYLEAADTPGWYCGPNFYTTEVLRKFARSQLDYILGKNPLKMSYVV 364
QY+VNAAFLA+LYSDYL+AADTPGWYCGPNFY+T VLR FARSQ+DYILGKNP KMSYVV
Sbjct: 443 QYSVNAAFLATLYSDYLDAADTPGWYCGPNFYSTSVLRDFARSQIDYILGKNPRKMSYVV 502
Query: 365 GFGNKYPKRPHHRGASIPHNGVKYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDV 544
GFG KYP+ HHRGASIP N VKY CKGG+KWRD+KK NPN + GAMVAGPD+RDGY+DV
Sbjct: 503 GFGTKYPRHVHHRGASIPKNKVKYNCKGGWKWRDSKKPNPNTIEGAMVAGPDKRDGYRDV 562
Query: 545 RTNYNYTEPTLAANAGLVAALISISDIKTGRFGIDKNTIFSAI 673
R NYNYTEPTLA NAGLVAAL+++S + IDKNTIFSA+
Sbjct: 563 RMNYNYTEPTLAGNAGLVAALVALSGEEEATGKIDKNTIFSAV 605
>sptr|Q94C24|Q94C24 AT5g49720/K2I5_8.
Length = 621
Score = 373 bits (958), Expect = e-102
Identities = 175/223 (78%), Positives = 195/223 (87%)
Frame = +2
Query: 5 EVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPLFNSFNFTKGGLIQLNHGRPQPL 184
++LLSRLRLFLSPG PYEEILRTFHNQT VMCSYLP+FN FN T GGLI+LNHG PQPL
Sbjct: 383 QLLLSRLRLFLSPGCPYEEILRTFHNQTSIVMCSYLPIFNKFNRTNGGLIELNHGAPQPL 442
Query: 185 QYAVNAAFLASLYSDYLEAADTPGWYCGPNFYTTEVLRKFARSQLDYILGKNPLKMSYVV 364
QY+VNAAFLA+LYSDYL+AADTPGWYCGPNFY+T VLR FARSQ+DYILGKNP KMSYVV
Sbjct: 443 QYSVNAAFLATLYSDYLDAADTPGWYCGPNFYSTSVLRDFARSQIDYILGKNPRKMSYVV 502
Query: 365 GFGNKYPKRPHHRGASIPHNGVKYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDV 544
GFG KYP+ HHRGASIP N VKY CKGG+KWRD+KK NPN + GAMVAGPD+RDGY+DV
Sbjct: 503 GFGTKYPRHVHHRGASIPKNKVKYNCKGGWKWRDSKKPNPNTIEGAMVAGPDKRDGYRDV 562
Query: 545 RTNYNYTEPTLAANAGLVAALISISDIKTGRFGIDKNTIFSAI 673
R NYNYTEPTLA NAGLVAAL+++S + IDKNTIFSA+
Sbjct: 563 RMNYNYTEPTLAGNAGLVAALVALSGEEEATGKIDKNTIFSAV 605
>sptr|Q9STW8|Q9STW8 Endo-1, 4-beta-glucanase like protein.
Length = 620
Score = 343 bits (880), Expect = 3e-93
Identities = 161/223 (72%), Positives = 188/223 (84%)
Frame = +2
Query: 5 EVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPLFNSFNFTKGGLIQLNHGRPQPL 184
++LL+R+RLFLSPGYPYE++L FHNQT VMCSYLP + FN T GGLIQLNHG PQPL
Sbjct: 384 QLLLTRMRLFLSPGYPYEDMLSEFHNQTGRVMCSYLPYYKKFNRTNGGLIQLNHGAPQPL 443
Query: 185 QYAVNAAFLASLYSDYLEAADTPGWYCGPNFYTTEVLRKFARSQLDYILGKNPLKMSYVV 364
QY NAAFLA+L+SDYLEAADTPGWYCGPNFYTTE LR F+RSQ+DYILGKNP KMSYVV
Sbjct: 444 QYVANAAFLAALFSDYLEAADTPGWYCGPNFYTTEFLRNFSRSQIDYILGKNPRKMSYVV 503
Query: 365 GFGNKYPKRPHHRGASIPHNGVKYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDV 544
G+G +YPK+ HHRGASIP N +K C GG+KW+ +KK NPN + GAMVAGPD+ DG+ D+
Sbjct: 504 GYGQRYPKQVHHRGASIPKN-MKETCTGGFKWKKSKKNNPNAINGAMVAGPDKHDGFHDI 562
Query: 545 RTNYNYTEPTLAANAGLVAALISISDIKTGRFGIDKNTIFSAI 673
RTNYNYTEPTLA NAGLVAAL+++S K GIDKNT+FSA+
Sbjct: 563 RTNYNYTEPTLAGNAGLVAALVALSGEKAVG-GIDKNTMFSAV 604
>sptr|Q7XUK4|Q7XUK4 OSJNBa0067K08.14 protein.
Length = 623
Score = 313 bits (801), Expect = 4e-84
Identities = 144/225 (64%), Positives = 175/225 (77%), Gaps = 2/225 (0%)
Frame = +2
Query: 5 EVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPLFNSFNFTKGGLIQLNHGRPQPL 184
E+LLSRLR+FL+PGYPYEE L +HN T MC+Y P F +FNFTKGGL Q NHG+ QPL
Sbjct: 383 ELLLSRLRMFLNPGYPYEESLIGYHNTTSMNMCTYFPRFGAFNFTKGGLAQFNHGKGQPL 442
Query: 185 QYAVNAAFLASLYSDYLEAADTPGWYCGPNFYTTEVLRKFARSQLDYILGKNPLKMSYVV 364
QY V +FLA+LY+DY+E+ + PGWYCGP F T + LR FARSQ++YILG NP KMSYVV
Sbjct: 443 QYTVANSFLAALYADYMESVNVPGWYCGPYFMTVDDLRSFARSQVNYILGDNPKKMSYVV 502
Query: 365 GFGNKYPKRPHHRGASIPHNGVKYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDV 544
G+G KYP+R HHRGAS PHNG+KY C GGYKWRDTK A+PN+LVGAMV GPD+ D +KD
Sbjct: 503 GYGKKYPRRLHHRGASTPHNGIKYSCTGGYKWRDTKGADPNVLVGAMVGGPDKNDQFKDA 562
Query: 545 RTNYNYTEPTLAANAGLVAALISISDIKTGR--FGIDKNTIFSAI 673
R Y EPTL NAGLVAAL+++++ G +DKNT+FSA+
Sbjct: 563 RLTYAQNEPTLVGNAGLVAALVALTNSGRGAGVTAVDKNTMFSAV 607
>sptr|O04478|O04478 F5I14.14.
Length = 623
Score = 276 bits (705), Expect = 5e-73
Identities = 126/222 (56%), Positives = 170/222 (76%)
Frame = +2
Query: 8 VLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPLFNSFNFTKGGLIQLNHGRPQPLQ 187
+L++R RLFL+PG+PYE +L +HN T MC+YL +N FN T GGL+QLN G+P+PL+
Sbjct: 388 LLMTRYRLFLNPGFPYENMLNRYHNATGITMCAYLKQYNVFNRTSGGLMQLNLGKPRPLE 447
Query: 188 YAVNAAFLASLYSDYLEAADTPGWYCGPNFYTTEVLRKFARSQLDYILGKNPLKMSYVVG 367
Y +A+FLASL++DYL + PGWYCGP F VL+ FA+SQ+DYILG NPLKMSYVVG
Sbjct: 448 YVAHASFLASLFADYLNSTGVPGWYCGPTFVENHVLKDFAQSQIDYILGDNPLKMSYVVG 507
Query: 368 FGNKYPKRPHHRGASIPHNGVKYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDVR 547
FG K+P+R HHRGA+IP++ + C+ G K+RDTK NPN + GAMV GP++ D + D+R
Sbjct: 508 FGKKFPRRVHHRGATIPNDKKRRSCREGLKYRDTKNPNPNNITGAMVGGPNKFDEFHDLR 567
Query: 548 TNYNYTEPTLAANAGLVAALISISDIKTGRFGIDKNTIFSAI 673
NYN +EPTL+ NAGLVAAL+S++ +G IDKNT+F+++
Sbjct: 568 NNYNASEPTLSGNAGLVAALVSLT--SSGGQQIDKNTMFNSV 607
>sptrnew|AAQ63883|AAQ63883 Cellulase.
Length = 601
Score = 198 bits (503), Expect = 1e-49
Identities = 97/206 (47%), Positives = 140/206 (67%), Gaps = 1/206 (0%)
Frame = +2
Query: 8 VLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPLFNSF-NFTKGGLIQLNHGRPQPL 184
VLL+ +R F PG+PYE++L+ N T ++MCSYL F + + T GGL+ L
Sbjct: 382 VLLTGIRYFRDPGFPYEDVLKFSSNSTHSLMCSYL--FKKYMSRTPGGLVIPKPDNGPLL 439
Query: 185 QYAVNAAFLASLYSDYLEAADTPGWYCGPNFYTTEVLRKFARSQLDYILGKNPLKMSYVV 364
QYAV A+FL+ LYSDY++ G C + ++ +LR F+ SQ++YILG+NP+KMSY+V
Sbjct: 440 QYAVTASFLSKLYSDYIDHLKISGASCETDTFSVSMLRDFSSSQVNYILGQNPMKMSYLV 499
Query: 365 GFGNKYPKRPHHRGASIPHNGVKYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDV 544
G+G+K+P + HHR ASIP + Y C G W ++K NP +L+GAMV GPD D + D
Sbjct: 500 GYGDKFPVQVHHRSASIPWDKRLYNCDDGKTWLNSKNPNPQVLLGAMVGGPDTNDHFTDQ 559
Query: 545 RTNYNYTEPTLAANAGLVAALISISD 622
R+N +TEPT+++NAGLV+ALI++ D
Sbjct: 560 RSNKRFTEPTISSNAGLVSALIALQD 585
>sptr|Q9FXI9|Q9FXI9 F6F9.1 protein (At1g19940/F6F9_1).
Length = 515
Score = 152 bits (383), Expect = 1e-35
Identities = 89/204 (43%), Positives = 119/204 (58%), Gaps = 2/204 (0%)
Frame = +2
Query: 8 VLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPLFNSFNF--TKGGLIQLNHGRPQP 181
+LLSRL F G + L+ F + VMC +P + T GGLI ++
Sbjct: 305 ILLSRLTFF-KKGLSGSKGLQGFKETAEAVMCGLIPSSPTATSSRTDGGLIWVSEWNA-- 361
Query: 182 LQYAVNAAFLASLYSDYLEAADTPGWYCGPNFYTTEVLRKFARSQLDYILGKNPLKMSYV 361
LQ+ V++AFLA+LYSDY+ + C + LRKFARSQ DY+LGKNP KMSY+
Sbjct: 362 LQHPVSSAFLATLYSDYMLTSGVKELSCSDQSFKPSDLRKFARSQADYMLGKNPEKMSYL 421
Query: 362 VGFGNKYPKRPHHRGASIPHNGVKYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKD 541
VG+G KYP+ HHRGASIP + GCK G+KW ++ + NPN+ GA+V GP D + D
Sbjct: 422 VGYGEKYPEFVHHRGASIPADATT-GCKDGFKWLNSDEPNPNVAYGALVGGPFLNDTFID 480
Query: 542 VRTNYNYTEPTLAANAGLVAALIS 613
R N EP+ N+ LV L+S
Sbjct: 481 ARNNSMQNEPS-TYNSALVVGLLS 503
>sw|P05522|GUN1_PERAE Endoglucanase 1 precursor (EC 3.2.1.4)
(Endo-1,4-beta-glucanase) (Abscission cellulase 1).
Length = 494
Score = 149 bits (376), Expect = 7e-35
Identities = 85/201 (42%), Positives = 117/201 (58%), Gaps = 7/201 (3%)
Frame = +2
Query: 26 RLFLSPGYPYEEI--LRTFHNQTDNVMCSYLPLFNSFN--FTKGGLIQLNHGRPQPLQYA 193
++ LS G+ + I L+ + TDN +CS +P +SF +T GGL L G LQY
Sbjct: 287 KVLLSKGFLQDRIEELQLYKVHTDNYICSLIPGTSSFQAQYTPGGL--LYKGSASNLQYV 344
Query: 194 VNAAFLASLYSDYLEAADTPGWYCGPNFYTTEVLRKFARSQLDYILGKNPLKMSYVVGFG 373
+ AFL Y++YL ++ CG T + L A+ Q+DYILG+NP KMSY+VGFG
Sbjct: 345 TSTAFLLLTYANYLNSSGGHA-SCGTTTVTAKNLISLAKKQVDYILGQNPAKMSYMVGFG 403
Query: 374 NKYPKRPHHRGASIPHNGV---KYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDV 544
+YP+ HHRG+S+P V C G+++ + NPNILVGA++ GPD RD + D
Sbjct: 404 ERYPQHVHHRGSSLPSVQVHPNSIPCNAGFQYLYSSPPNPNILVGAILGGPDNRDSFSDD 463
Query: 545 RTNYNYTEPTLAANAGLVAAL 607
R NY +EP NA LV AL
Sbjct: 464 RNNYQQSEPATYINAPLVGAL 484
>sptr|Q43149|Q43149 Cellulase (EC 3.2.1.4) (Fragment).
Length = 508
Score = 147 bits (370), Expect = 4e-34
Identities = 82/197 (41%), Positives = 119/197 (60%), Gaps = 9/197 (4%)
Frame = +2
Query: 65 LRTFHNQTDNVMCSYLPLFNSFNF--TKGGLIQLNHGRPQPLQYAVNAAFLASLYSDYLE 238
L F + +++ +C+ +P +S T GGL+ LQY +A+ + +YS L
Sbjct: 298 LGKFKSDSESFICALMPGSSSVQIQTTPGGLLYTRDS--SNLQYVTSASMVFLIYSKILN 355
Query: 239 AADTPGWYCGPNFYTTEVLRKFARSQLDYILGKNPLKMSYVVGFGNKYPKRPHHRGASIP 418
AA+ G CG + T ++ FA+SQ+DYILG NP+KMSY+VGFG+KYP + HHRGASIP
Sbjct: 356 AANIKGVQCGSVNFPTSQIKAFAKSQVDYILGNNPMKMSYMVGFGSKYPLQLHHRGASIP 415
Query: 419 H---NGVKYGCKGGYK-WRDTKKANPNILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAAN 586
+ + GC GY W + K+NPN VGA+V GP+ D + DVR++Y++ E T N
Sbjct: 416 SMQIHPARVGCNEGYSAWYSSSKSNPNTHVGAIVGGPNSNDQFNDVRSDYSHLETTTYMN 475
Query: 587 A---GLVAALISISDIK 628
A G VAAL+ + I+
Sbjct: 476 AAFVGSVAALVGDNSIR 492
>sptr|Q9LR07|Q9LR07 F10A5.13 (Hypothetical protein) (Putative
endo-beta-1,4-glucanase).
Length = 525
Score = 145 bits (367), Expect = 8e-34
Identities = 89/206 (43%), Positives = 119/206 (57%), Gaps = 2/206 (0%)
Frame = +2
Query: 5 EVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPLF--NSFNFTKGGLIQLNHGRPQ 178
+VLLSRL LF + L + N VMC LP ++ + T GGLI ++
Sbjct: 313 QVLLSRL-LFFKKDLSGSKGLGNYRNTAKAVMCGLLPKSPTSTASRTNGGLIWVSEWNS- 370
Query: 179 PLQYAVNAAFLASLYSDYLEAADTPGWYCGPNFYTTEVLRKFARSQLDYILGKNPLKMSY 358
+Q +V++AFLASL+SDY+ + C + LR FA+SQ DY+LGKNPL S+
Sbjct: 371 -MQQSVSSAFLASLFSDYMLTSRIHKISCDGKIFKATELRDFAKSQADYMLGKNPLGTSF 429
Query: 359 VVGFGNKYPKRPHHRGASIPHNGVKYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYK 538
VVG+G+KYP+ HHRGASIP + GC G+KW ++ K NPNI GA+V GP + +
Sbjct: 430 VVGYGDKYPQFVHHRGASIPADATT-GCLDGFKWFNSTKPNPNIAYGALVGGPFFNETFT 488
Query: 539 DVRTNYNYTEPTLAANAGLVAALISI 616
D R N EPT NA LV L S+
Sbjct: 489 DSRENPMQNEPTTYNNALLVGLLSSL 514
>sptr|Q8LCP6|Q8LCP6 Endo-beta-1,4-glucanase, putative.
Length = 525
Score = 145 bits (367), Expect = 8e-34
Identities = 89/206 (43%), Positives = 119/206 (57%), Gaps = 2/206 (0%)
Frame = +2
Query: 5 EVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPLF--NSFNFTKGGLIQLNHGRPQ 178
+VLLSRL LF + L + N VMC LP ++ + T GGLI ++
Sbjct: 313 QVLLSRL-LFFKKDLSGSKGLGNYRNTAKAVMCGLLPKSPTSTASRTNGGLIWVSEWNS- 370
Query: 179 PLQYAVNAAFLASLYSDYLEAADTPGWYCGPNFYTTEVLRKFARSQLDYILGKNPLKMSY 358
+Q +V++AFLASL+SDY+ + C + LR FA+SQ DY+LGKNPL S+
Sbjct: 371 -MQQSVSSAFLASLFSDYMLTSRIHKISCDGKIFKATELRDFAKSQADYMLGKNPLGTSF 429
Query: 359 VVGFGNKYPKRPHHRGASIPHNGVKYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYK 538
VVG+G+KYP+ HHRGASIP + GC G+KW ++ K NPNI GA+V GP + +
Sbjct: 430 VVGYGDKYPQFVHHRGASIPADATT-GCLDGFKWFNSTKPNPNIAYGALVGGPFFNETFT 488
Query: 539 DVRTNYNYTEPTLAANAGLVAALISI 616
D R N EPT NA LV L S+
Sbjct: 489 DSRENPMQNEPTTYNNALLVGLLSSL 514
>sptr|Q9SUS0|Q9SUS0 Putative cellulase.
Length = 479
Score = 145 bits (366), Expect = 1e-33
Identities = 83/205 (40%), Positives = 115/205 (56%), Gaps = 8/205 (3%)
Frame = +2
Query: 29 LFLSPGYPYEEILRTFHNQTDNVMCSYLPLFNSFNF--TKGGLIQLNHGRPQPLQYAVNA 202
L +S Y L F + ++ +C+ +P +S T GGL+ + LQY A
Sbjct: 276 LLVSEFYNGANDLAKFKSDVESFVCAMMPGSSSQQIKPTPGGLLFIRDS--SNLQYVTTA 333
Query: 203 AFLASLYSDYLEAADTPGWYCGPNFYTTEVLRKFARSQLDYILGKNPLKMSYVVGFGNKY 382
+ YS L A CG +T +R FA+SQ+DYILG NP+KMSY+VGFG KY
Sbjct: 334 TTVLFHYSKTLTKAGVGSIQCGSTKFTVSQIRNFAKSQVDYILGNNPMKMSYMVGFGTKY 393
Query: 383 PKRPHHRGASIP---HNGVKYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDVRTN 553
P +PHHRG+S+P K C GGY + ++ NPN+ +GA+V GP+ D Y D +++
Sbjct: 394 PTQPHHRGSSLPSIQSKPEKIDCNGGYSYYNSDTPNPNVHIGAIVGGPNSSDQYSDKKSD 453
Query: 554 YNYTEPTLAANA---GLVAALISIS 619
Y++ EPT NA G VAALIS S
Sbjct: 454 YSHAEPTTYINAAFIGPVAALISSS 478
>sptr|Q9SRX3|Q9SRX3 Endo-1,4-beta glucanase.
Length = 501
Score = 145 bits (365), Expect = 1e-33
Identities = 82/199 (41%), Positives = 117/199 (58%), Gaps = 5/199 (2%)
Frame = +2
Query: 26 RLFLSPGYPYEEI--LRTFHNQTDNVMCSYLPLFNSFNFTKGGLIQLNHGRPQPLQYAVN 199
R+ LS + +++ L + D+ +CS LP +S +T GGL+ G +QY +
Sbjct: 300 RILLSKEFLIQKVKSLEEYKEHADSFICSVLPGASSSQYTPGGLL-FKMGESN-MQYVTS 357
Query: 200 AAFLASLYSDYLEAADTPGWYCGPNFYTTEVLRKFARSQLDYILGKNPLKMSYVVGFGNK 379
+FL Y+ YL +A T YCG + T LR A+ Q+DY+LG NPLKMSY+VG+G K
Sbjct: 358 TSFLLLTYAKYLTSARTVA-YCGGSVVTPARLRSIAKKQVDYLLGGNPLKMSYMVGYGLK 416
Query: 380 YPKRPHHRGASIPHNGV---KYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDVRT 550
YP+R HHRG+S+P V + C G+ ++ NPN LVGA+V GPD+ D + D R+
Sbjct: 417 YPRRIHHRGSSLPSVAVHPTRIQCHDGFSLFTSQSPNPNDLVGAVVGGPDQNDQFPDERS 476
Query: 551 NYNYTEPTLAANAGLVAAL 607
+Y +EP NA LV AL
Sbjct: 477 DYGRSEPATYINAPLVGAL 495
>sptr|Q9SZ90|Q9SZ90 Cellulase-like protein.
Length = 480
Score = 145 bits (365), Expect = 1e-33
Identities = 84/205 (40%), Positives = 111/205 (54%), Gaps = 8/205 (3%)
Frame = +2
Query: 29 LFLSPGYPYEEILRTFHNQTDNVMCSYLPLFNSFNF--TKGGLIQLNHGRPQPLQYAVNA 202
L S Y L F ++ +C+ +P +S T GG++ + LQY A
Sbjct: 278 LLASEFYNGANDLEKFKTDVESFVCALMPGSSSQQIKPTPGGILFIRDS--SNLQYVTTA 335
Query: 203 AFLASLYSDYLEAADTPGWYCGPNFYTTEVLRKFARSQLDYILGKNPLKMSYVVGFGNKY 382
+ YS L A CG +T +R FA+SQ+DYILG NPLKMSY+VGFG KY
Sbjct: 336 TTILFYYSKTLTKAGVGSIQCGSTQFTVSQIRNFAKSQVDYILGNNPLKMSYMVGFGTKY 395
Query: 383 PKRPHHRGASIP---HNGVKYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDVRTN 553
P +PHHRG+S+P K C GG+ + + NPN+ GA+V GP+ D Y D RT+
Sbjct: 396 PTQPHHRGSSLPSIQSKPEKIDCNGGFSYYNFDTPNPNVHTGAIVGGPNSSDQYSDKRTD 455
Query: 554 YNYTEPTLAANA---GLVAALISIS 619
Y++ EPT NA G VAALIS S
Sbjct: 456 YSHAEPTTYINAAFIGSVAALISSS 480
>sptr|O64949|O64949 Endo-1,4-beta glucanase.
Length = 501
Score = 144 bits (363), Expect = 2e-33
Identities = 81/199 (40%), Positives = 117/199 (58%), Gaps = 5/199 (2%)
Frame = +2
Query: 26 RLFLSPGYPYEEI--LRTFHNQTDNVMCSYLPLFNSFNFTKGGLIQLNHGRPQPLQYAVN 199
R+ LS + +++ L + D+ +CS +P +S +T GGL+ G +QY +
Sbjct: 300 RILLSKEFLIQKVKSLEEYKEHADSFICSVIPGASSSQYTPGGLL-FKMGESN-MQYVTS 357
Query: 200 AAFLASLYSDYLEAADTPGWYCGPNFYTTEVLRKFARSQLDYILGKNPLKMSYVVGFGNK 379
+FL Y+ YL +A T YCG + T LR A+ Q+DY+LG NPLKMSY+VG+G K
Sbjct: 358 TSFLLLTYAKYLTSARTVA-YCGGSVVTPARLRSIAKKQVDYLLGGNPLKMSYMVGYGLK 416
Query: 380 YPKRPHHRGASIPHNGV---KYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDVRT 550
YP+R HHRG+S+P V + C G+ ++ NPN LVGA+V GPD+ D + D R+
Sbjct: 417 YPRRIHHRGSSLPSVAVHPTRIQCHDGFSLFTSQSPNPNDLVGAVVGGPDQNDQFPDERS 476
Query: 551 NYNYTEPTLAANAGLVAAL 607
+Y +EP NA LV AL
Sbjct: 477 DYGRSEPATYINAPLVGAL 495
>sptr|O82473|O82473 Endo-1,4-beta-glucanase (EC 3.2.1.4).
Length = 497
Score = 144 bits (362), Expect = 3e-33
Identities = 80/184 (43%), Positives = 112/184 (60%), Gaps = 6/184 (3%)
Frame = +2
Query: 86 TDNVMCSYLPLFNSFN--FTKGGLIQLNHGRPQPLQYAVNAAFLASLYSDYLEAADTPGW 259
+DN +CS +P SF +T GGL+ G LQY +++FL Y+ YL + +
Sbjct: 315 SDNYICSLIPGSPSFQAQYTPGGLLYKGSG--SNLQYVTSSSFLLLTYAKYLRS-NGGDV 371
Query: 260 YCGPNFYTTEVLRKFARSQLDYILGKNPLKMSYVVGFGNKYPKRPHHRGASIP----HNG 427
CG + + E L + A+ Q+DYILG NP K+SY+VGFG+KYP R HHRG+S+P H G
Sbjct: 372 SCGSSRFPAERLVELAKKQVDYILGDNPAKISYMVGFGDKYPLRVHHRGSSLPSVHAHPG 431
Query: 428 VKYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAANAGLVAAL 607
GC G++ ++ NPN+L+GA+V GPD RD ++D R NY +EP NA LV AL
Sbjct: 432 -HIGCNDGFQSLNSGSPNPNVLIGAIVGGPDSRDNFEDDRNNYQQSEPATYINAPLVGAL 490
Query: 608 ISIS 619
+S
Sbjct: 491 AFLS 494
>sptr|O81416|O81416 T2H3.5 protein (Putative endo-1, 4-beta
glucanase) (AT4g02290/T2H3_5).
Length = 516
Score = 142 bits (357), Expect = 1e-32
Identities = 79/201 (39%), Positives = 118/201 (58%), Gaps = 7/201 (3%)
Frame = +2
Query: 26 RLFLSPGYPYEEI--LRTFHNQTDNVMCSYLP--LFNSFNFTKGGLIQLNHGRPQPLQYA 193
R+ L+ + + + L + DN +CS +P F+S +T GGL L +QY
Sbjct: 309 RILLTKAFLVQNVKTLHEYKGHADNFICSVIPGAPFSSTQYTPGGL--LFKMADANMQYV 366
Query: 194 VNAAFLASLYSDYLEAADTPGWYCGPNFYTTEVLRKFARSQLDYILGKNPLKMSYVVGFG 373
+ +FL Y+ YL +A T +CG + YT LR A+ Q+DY+LG NPL+MSY+VG+G
Sbjct: 367 TSTSFLLLTYAKYLTSAKTVV-HCGGSVYTPGRLRSIAKRQVDYLLGDNPLRMSYMVGYG 425
Query: 374 NKYPKRPHHRGASIP---HNGVKYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDV 544
K+P+R HHRG+S+P + K C G+ +++ NPN LVGA+V GPD+ D + D
Sbjct: 426 PKFPRRIHHRGSSLPCVASHPAKIQCHQGFAIMNSQSPNPNFLVGAVVGGPDQHDRFPDE 485
Query: 545 RTNYNYTEPTLAANAGLVAAL 607
R++Y +EP N+ LV AL
Sbjct: 486 RSDYEQSEPATYINSPLVGAL 506
>sptr|Q42871|Q42871 Endo-1,4-beta-glucanase precursor (EC 3.2.1.4).
Length = 501
Score = 142 bits (357), Expect = 1e-32
Identities = 75/192 (39%), Positives = 111/192 (57%), Gaps = 6/192 (3%)
Frame = +2
Query: 65 LRTFHNQTDNVMCSYLPLFNSFNF--TKGGLIQLNHGRPQPLQYAVNAAFLASLYSDYLE 238
L F D+ +C +P +S T GGL+ LQY A + +Y+ LE
Sbjct: 291 LEKFKKDADSFICGLMPESSSIQIKTTPGGLLYYRDS--SNLQYVNGATMVLFMYTKVLE 348
Query: 239 AADTPGWYCGPNFYTTEVLRKFARSQLDYILGKNPLKMSYVVGFGNKYPKRPHHRGASIP 418
AA G CG ++T ++ FA+ Q+DYILG NPLKMSY+VGFGNKYP + HHR +S+P
Sbjct: 349 AAGIGGVTCGSVNFSTSKIKAFAKLQVDYILGNNPLKMSYMVGFGNKYPTKLHHRASSLP 408
Query: 419 ---HNGVKYGCKGGY-KWRDTKKANPNILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAAN 586
++ + GC GY W ++ NPN VGA+V GP+ D + D R++Y+++EPT N
Sbjct: 409 SIYNHPTRVGCNDGYSSWYNSNNPNPNTHVGAIVGGPNSGDQFIDSRSDYSHSEPTTYMN 468
Query: 587 AGLVAALISISD 622
A + ++ ++ D
Sbjct: 469 AAFIGSVAALID 480
>sptr|Q9FVQ2|Q9FVQ2 Endo-beta-1,4-glucanase, putative.
Length = 623
Score = 141 bits (355), Expect = 2e-32
Identities = 87/219 (39%), Positives = 116/219 (52%), Gaps = 6/219 (2%)
Frame = +2
Query: 5 EVLLSRLRLFLSPGYPYEEILRTFHNQTDNVMCSYLPLFNSFNF--TKGGLIQLNHGRPQ 178
++LLS++ L G Y L+ + + D C+ L +N T GGL+ +
Sbjct: 284 QILLSKI-LLEGKGGIYTSTLKQYQTKADYFACACLKKNGGYNIQTTPGGLMYVREWNN- 341
Query: 179 PLQYAVNAAFLASLYSDYLEAADTPGWYCGPNFYTTEVLRKFARSQLDYILGKNPLKMSY 358
LQYA AA+L ++YSDYL AA+ C + L FARSQ DYILGKN MSY
Sbjct: 342 -LQYASAAAYLLAVYSDYLSAANAK-LNCPDGLVQPQGLLDFARSQADYILGKNRQGMSY 399
Query: 359 VVGFGNKYPKRPHHRGASIPH---NGVKYGCKGGY-KWRDTKKANPNILVGAMVAGPDRR 526
VVG+G KYP R HHRG+SIP C G+ W + +PN++ GA+V GPD
Sbjct: 400 VVGYGPKYPIRVHHRGSSIPSIFAQRSSVSCVQGFDSWYRRSQGDPNVIYGALVGGPDEN 459
Query: 527 DGYKDVRTNYNYTEPTLAANAGLVAALISISDIKTGRFG 643
D Y D R+NY +EPTL+ A LV + G +G
Sbjct: 460 DNYSDDRSNYEQSEPTLSGTAPLVGLFAKLYGGSLGSYG 498
>sptr|Q93WZ1|Q93WZ1 Endo-beta-1,4-glucanase precursor (EC 3.2.1.4).
Length = 489
Score = 141 bits (355), Expect = 2e-32
Identities = 76/184 (41%), Positives = 112/184 (60%), Gaps = 6/184 (3%)
Frame = +2
Query: 74 FHNQTDNVMCSYLPL--FNSFNFTKGGLIQLNHGRPQPLQYAVNAAFLASLYSDYLEAAD 247
F Q ++ +C LP + S +TKGGLI + LQY + L + Y+ Y+ A+
Sbjct: 301 FRQQAEDFVCKILPNSPYTSTQYTKGGLIYKL--TEENLQYVTSITSLLTTYAKYM-ASK 357
Query: 248 TPGWYCGPNFYTTEVLRKFARSQLDYILGKNPLKMSYVVGFGNKYPKRPHHRGASIPHNG 427
+ CG T + +R A+ Q+DYILG NP+KMSY+VG+G YP+R HHRG+S+P
Sbjct: 358 KHTFNCGSLLVTEKTIRILAKRQVDYILGNNPMKMSYMVGYGTNYPRRVHHRGSSLPSMA 417
Query: 428 V---KYGCKGGYK-WRDTKKANPNILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAANAGL 595
+ +GC GG++ + T ANPNILVGA+V GP++ D + D RT+Y+++EP NA +
Sbjct: 418 MHPQSFGCDGGFQPYYYTANANPNILVGAIVGGPNQNDFFPDERTDYSHSEPATYINAAI 477
Query: 596 VAAL 607
V L
Sbjct: 478 VGPL 481
Database: /db/uniprot/tmp/swall
Posted date: Mar 5, 2004 7:45 PM
Number of letters in database: 442,889,342
Number of sequences in database: 1,395,590
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 831,249,840
Number of Sequences: 1395590
Number of extensions: 17026241
Number of successful extensions: 40480
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 38427
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 40189
length of database: 442,889,342
effective HSP length: 124
effective length of database: 269,836,182
effective search space used: 63141666588
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

