BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= 2161200.2.1
(845 letters)
Database: /db/uniprot/tmp/swall
1,395,590 sequences; 442,889,342 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
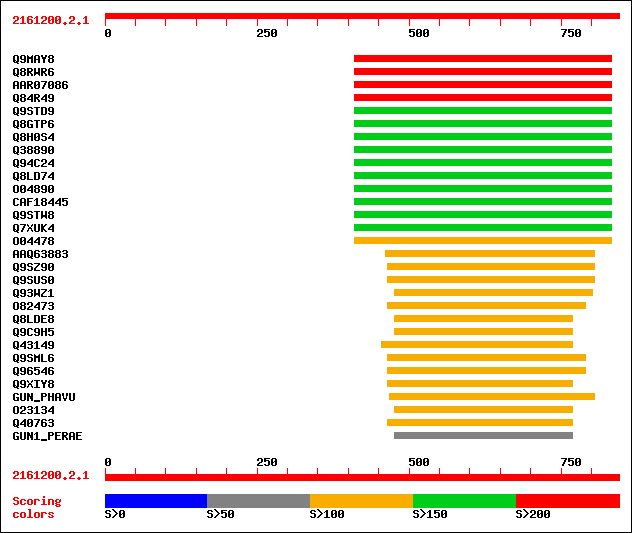
sptr|Q9MAY8|Q9MAY8 Endo-1,4-beta-glucanase Cel1. 230 2e-59
sptr|Q8RWR6|Q8RWR6 Endo-1,4-beta-glucanase. 229 5e-59
sptrnew|AAR07086|AAR07086 Putative endo-1,4-beta-glucanase. 223 3e-57
sptr|Q84R49|Q84R49 Putative endo-1,4-beta-glucanase. 223 3e-57
sptr|Q9STD9|Q9STD9 Cellulase (EC 3.2.1.4). 194 1e-48
sptr|Q8GTP6|Q8GTP6 Endo-1,4-beta-D-glucanase (EC 3.2.1.4). 192 4e-48
sptr|Q8H0S4|Q8H0S4 At5g49720/K2I5_8. 192 7e-48
sptr|Q38890|Q38890 Cellulase precursor (Cellulase homolog OR16PE... 192 7e-48
sptr|Q94C24|Q94C24 AT5g49720/K2I5_8. 192 7e-48
sptr|Q8LD74|Q8LD74 Cellulase homolog OR16pep. 192 7e-48
sptr|O04890|O04890 Endo-1,4-beta-glucanase (EC 3.2.1.4). 190 3e-47
sptrnew|CAF18445|CAF18445 Endo-1,4-beta-D-glucanase KORRIGAN (EC... 189 6e-47
sptr|Q9STW8|Q9STW8 Endo-1, 4-beta-glucanase like protein. 171 1e-41
sptr|Q7XUK4|Q7XUK4 OSJNBa0067K08.14 protein. 167 1e-40
sptr|O04478|O04478 F5I14.14. 145 7e-34
sptrnew|AAQ63883|AAQ63883 Cellulase. 121 1e-26
sptr|Q9SZ90|Q9SZ90 Cellulase-like protein. 109 4e-23
sptr|Q9SUS0|Q9SUS0 Putative cellulase. 109 4e-23
sptr|Q93WZ1|Q93WZ1 Endo-beta-1,4-glucanase precursor (EC 3.2.1.4). 105 8e-22
sptr|O82473|O82473 Endo-1,4-beta-glucanase (EC 3.2.1.4). 104 1e-21
sptr|Q8LDE8|Q8LDE8 Putative beta-glucanase. 103 2e-21
sptr|Q9C9H5|Q9C9H5 Putative beta-glucanase, 74324-76084. 103 2e-21
sptr|Q43149|Q43149 Cellulase (EC 3.2.1.4) (Fragment). 103 4e-21
sptr|Q9SML6|Q9SML6 Endo-beta-1,4-glucanase precursor (EC 3.2.1.4). 103 4e-21
sptr|Q96546|Q96546 Endo-beta-1,4-glucanase (EC 3.2.1.4). 103 4e-21
sptr|Q9XIY8|Q9XIY8 Endo-1,4-beta glucanase precursor. 102 5e-21
sw|P22503|GUN_PHAVU Endoglucanase precursor (EC 3.2.1.4) (Endo-1... 102 9e-21
sptr|O23134|O23134 Beta-glucanase. 102 9e-21
sptr|Q40763|Q40763 Cellulase precursor. 101 2e-20
sw|P05522|GUN1_PERAE Endoglucanase 1 precursor (EC 3.2.1.4) (End... 100 2e-20
>sptr|Q9MAY8|Q9MAY8 Endo-1,4-beta-glucanase Cel1.
Length = 621
Score = 230 bits (586), Expect = 2e-59
Identities = 108/141 (76%), Positives = 118/141 (83%)
Frame = -2
Query: 832 PXXYXXPXXYTIXVLXKXXXXQLDYIXXKNPLKMSYXVGFXNKYPKRPHHRGASIPHNGV 653
P Y P YT VL K QLDYI KNP KMSY VGF KYPKR HHRGASIPHNGV
Sbjct: 465 PGWYCGPNFYTTDVLRKFAKSQLDYILGKNPQKMSYVVGFGKKYPKRVHHRGASIPHNGV 524
Query: 652 KYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAANAGLVAALI 473
KYGCKGG+KWR++KKANPNILVGAMVAGPD+ DG+KD+RTNYNYTEPTLAANAGLVAALI
Sbjct: 525 KYGCKGGFKWRESKKANPNILVGAMVAGPDKHDGFKDIRTNYNYTEPTLAANAGLVAALI 584
Query: 472 SISDIKTGRFGIDKNTIFSAI 410
S++DI TGR+ IDKNTIFSA+
Sbjct: 585 SLADIDTGRYSIDKNTIFSAV 605
>sptr|Q8RWR6|Q8RWR6 Endo-1,4-beta-glucanase.
Length = 620
Score = 229 bits (583), Expect = 5e-59
Identities = 108/141 (76%), Positives = 117/141 (82%)
Frame = -2
Query: 832 PXXYXXPXXYTIXVLXKXXXXQLDYIXXKNPLKMSYXVGFXNKYPKRPHHRGASIPHNGV 653
P Y P YT VL K QLDYI KNP KMSY VGF KYPKR HHRGASIPHNGV
Sbjct: 465 PRWYCRPNFYTTDVLRKFAKSQLDYILGKNPQKMSYVVGFGKKYPKRVHHRGASIPHNGV 524
Query: 652 KYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAANAGLVAALI 473
KYGCKGG+KWR+ KKANPNILVGAMVAGPD+ DG+KD+RTNYNYTEPTLAANAGLVAALI
Sbjct: 525 KYGCKGGFKWREFKKANPNILVGAMVAGPDKHDGFKDIRTNYNYTEPTLAANAGLVAALI 584
Query: 472 SISDIKTGRFGIDKNTIFSAI 410
S++DI TGR+ IDKNTIFSA+
Sbjct: 585 SLADIDTGRYSIDKNTIFSAV 605
>sptrnew|AAR07086|AAR07086 Putative endo-1,4-beta-glucanase.
Length = 620
Score = 223 bits (568), Expect = 3e-57
Identities = 106/141 (75%), Positives = 117/141 (82%)
Frame = -2
Query: 832 PXXYXXPXXYTIXVLXKXXXXQLDYIXXKNPLKMSYXVGFXNKYPKRPHHRGASIPHNGV 653
P Y P YT VL K QLDY+ KNPLKMSY VGF NKYPKR HHRGASIPHNGV
Sbjct: 465 PGWYCGPTFYTTEVLRKFARSQLDYVLGKNPLKMSYVVGFGNKYPKRAHHRGASIPHNGV 524
Query: 652 KYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAANAGLVAALI 473
KYGCKGG+KWR+TKK NPNIL+GA+VAGPDR DG+KDVRTNYNYTEPTLAANAGLVAALI
Sbjct: 525 KYGCKGGFKWRETKKPNPNILIGALVAGPDRHDGFKDVRTNYNYTEPTLAANAGLVAALI 584
Query: 472 SISDIKTGRFGIDKNTIFSAI 410
S+++I + GIDKNTIFSA+
Sbjct: 585 SLTNIHV-KSGIDKNTIFSAV 604
>sptr|Q84R49|Q84R49 Putative endo-1,4-beta-glucanase.
Length = 620
Score = 223 bits (568), Expect = 3e-57
Identities = 106/141 (75%), Positives = 117/141 (82%)
Frame = -2
Query: 832 PXXYXXPXXYTIXVLXKXXXXQLDYIXXKNPLKMSYXVGFXNKYPKRPHHRGASIPHNGV 653
P Y P YT VL K QLDY+ KNPLKMSY VGF NKYPKR HHRGASIPHNGV
Sbjct: 465 PGWYCGPTFYTTEVLRKFARSQLDYVLGKNPLKMSYVVGFGNKYPKRAHHRGASIPHNGV 524
Query: 652 KYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAANAGLVAALI 473
KYGCKGG+KWR+TKK NPNIL+GA+VAGPDR DG+KDVRTNYNYTEPTLAANAGLVAALI
Sbjct: 525 KYGCKGGFKWRETKKPNPNILIGALVAGPDRHDGFKDVRTNYNYTEPTLAANAGLVAALI 584
Query: 472 SISDIKTGRFGIDKNTIFSAI 410
S+++I + GIDKNTIFSA+
Sbjct: 585 SLTNIHV-KSGIDKNTIFSAV 604
>sptr|Q9STD9|Q9STD9 Cellulase (EC 3.2.1.4).
Length = 621
Score = 194 bits (493), Expect = 1e-48
Identities = 92/141 (65%), Positives = 106/141 (75%)
Frame = -2
Query: 832 PXXYXXPXXYTIXVLXKXXXXQLDYIXXKNPLKMSYXVGFXNKYPKRPHHRGASIPHNGV 653
P Y P Y+ VL + Q+DYI KNP KMSY VGF KYPK HHRGASIP N V
Sbjct: 465 PGWYCGPNFYSTNVLREFARTQIDYILGKNPRKMSYLVGFGTKYPKHVHHRGASIPKNKV 524
Query: 652 KYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAANAGLVAALI 473
KY CKGG+KWRD+KK NPN + GAMVAGPD+RDG++DVRTNYNYTEPTLA NAGLVAAL+
Sbjct: 525 KYNCKGGWKWRDSKKPNPNTIEGAMVAGPDKRDGFRDVRTNYNYTEPTLAGNAGLVAALV 584
Query: 472 SISDIKTGRFGIDKNTIFSAI 410
++S + IDKNTIFSA+
Sbjct: 585 ALSGEEEASGTIDKNTIFSAV 605
>sptr|Q8GTP6|Q8GTP6 Endo-1,4-beta-D-glucanase (EC 3.2.1.4).
Length = 621
Score = 192 bits (489), Expect = 4e-48
Identities = 92/141 (65%), Positives = 107/141 (75%)
Frame = -2
Query: 832 PXXYXXPXXYTIXVLXKXXXXQLDYIXXKNPLKMSYXVGFXNKYPKRPHHRGASIPHNGV 653
P Y P Y+ VL + Q+DYI KNP KMSY VGF N YPK HHRGASIP N +
Sbjct: 467 PGWYCGPNFYSTDVLREFAKTQIDYILGKNPRKMSYVVGFGNHYPKHVHHRGASIPKNKI 526
Query: 652 KYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAANAGLVAALI 473
KY CKGG+KWRDT KANPN + GAMVAGPD+ DG++DVR+NYNYTEPTLA NAGLVAAL+
Sbjct: 527 KYSCKGGWKWRDTPKANPNTIDGAMVAGPDKHDGFRDVRSNYNYTEPTLAGNAGLVAALV 586
Query: 472 SISDIKTGRFGIDKNTIFSAI 410
++S K+ GIDKNTIFSA+
Sbjct: 587 ALSGEKS--VGIDKNTIFSAV 605
>sptr|Q8H0S4|Q8H0S4 At5g49720/K2I5_8.
Length = 621
Score = 192 bits (487), Expect = 7e-48
Identities = 91/141 (64%), Positives = 104/141 (73%)
Frame = -2
Query: 832 PXXYXXPXXYTIXVLXKXXXXQLDYIXXKNPLKMSYXVGFXNKYPKRPHHRGASIPHNGV 653
P Y P Y+ VL Q+DYI KNP KMSY VGF KYP+ HHRGASIP N V
Sbjct: 465 PGWYCGPNFYSTSVLRDFARSQIDYILGKNPRKMSYVVGFGTKYPRHVHHRGASIPKNKV 524
Query: 652 KYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAANAGLVAALI 473
KY CKGG+KWRD+KK NPN + GAMVAGPD+RDGY+DVR NYNYTEPTLA NAGLVAAL+
Sbjct: 525 KYNCKGGWKWRDSKKPNPNTIEGAMVAGPDKRDGYRDVRMNYNYTEPTLAGNAGLVAALV 584
Query: 472 SISDIKTGRFGIDKNTIFSAI 410
++S + IDKNTIFSA+
Sbjct: 585 ALSGEEEATGKIDKNTIFSAV 605
>sptr|Q38890|Q38890 Cellulase precursor (Cellulase homolog OR16PEP
precursor).
Length = 621
Score = 192 bits (487), Expect = 7e-48
Identities = 91/141 (64%), Positives = 104/141 (73%)
Frame = -2
Query: 832 PXXYXXPXXYTIXVLXKXXXXQLDYIXXKNPLKMSYXVGFXNKYPKRPHHRGASIPHNGV 653
P Y P Y+ VL Q+DYI KNP KMSY VGF KYP+ HHRGASIP N V
Sbjct: 465 PGWYCGPNFYSTSVLRDFARSQIDYILGKNPRKMSYVVGFGTKYPRHVHHRGASIPKNKV 524
Query: 652 KYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAANAGLVAALI 473
KY CKGG+KWRD+KK NPN + GAMVAGPD+RDGY+DVR NYNYTEPTLA NAGLVAAL+
Sbjct: 525 KYNCKGGWKWRDSKKPNPNTIEGAMVAGPDKRDGYRDVRMNYNYTEPTLAGNAGLVAALV 584
Query: 472 SISDIKTGRFGIDKNTIFSAI 410
++S + IDKNTIFSA+
Sbjct: 585 ALSGEEEATGKIDKNTIFSAV 605
>sptr|Q94C24|Q94C24 AT5g49720/K2I5_8.
Length = 621
Score = 192 bits (487), Expect = 7e-48
Identities = 91/141 (64%), Positives = 104/141 (73%)
Frame = -2
Query: 832 PXXYXXPXXYTIXVLXKXXXXQLDYIXXKNPLKMSYXVGFXNKYPKRPHHRGASIPHNGV 653
P Y P Y+ VL Q+DYI KNP KMSY VGF KYP+ HHRGASIP N V
Sbjct: 465 PGWYCGPNFYSTSVLRDFARSQIDYILGKNPRKMSYVVGFGTKYPRHVHHRGASIPKNKV 524
Query: 652 KYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAANAGLVAALI 473
KY CKGG+KWRD+KK NPN + GAMVAGPD+RDGY+DVR NYNYTEPTLA NAGLVAAL+
Sbjct: 525 KYNCKGGWKWRDSKKPNPNTIEGAMVAGPDKRDGYRDVRMNYNYTEPTLAGNAGLVAALV 584
Query: 472 SISDIKTGRFGIDKNTIFSAI 410
++S + IDKNTIFSA+
Sbjct: 585 ALSGEEEATGKIDKNTIFSAV 605
>sptr|Q8LD74|Q8LD74 Cellulase homolog OR16pep.
Length = 621
Score = 192 bits (487), Expect = 7e-48
Identities = 91/141 (64%), Positives = 104/141 (73%)
Frame = -2
Query: 832 PXXYXXPXXYTIXVLXKXXXXQLDYIXXKNPLKMSYXVGFXNKYPKRPHHRGASIPHNGV 653
P Y P Y+ VL Q+DYI KNP KMSY VGF KYP+ HHRGASIP N V
Sbjct: 465 PGWYCGPNFYSTSVLRDFARSQIDYILGKNPRKMSYVVGFGTKYPRHVHHRGASIPKNKV 524
Query: 652 KYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAANAGLVAALI 473
KY CKGG+KWRD+KK NPN + GAMVAGPD+RDGY+DVR NYNYTEPTLA NAGLVAAL+
Sbjct: 525 KYNCKGGWKWRDSKKPNPNTIEGAMVAGPDKRDGYRDVRMNYNYTEPTLAGNAGLVAALV 584
Query: 472 SISDIKTGRFGIDKNTIFSAI 410
++S + IDKNTIFSA+
Sbjct: 585 ALSGEEEATGKIDKNTIFSAV 605
>sptr|O04890|O04890 Endo-1,4-beta-glucanase (EC 3.2.1.4).
Length = 617
Score = 190 bits (482), Expect = 3e-47
Identities = 91/141 (64%), Positives = 107/141 (75%)
Frame = -2
Query: 832 PXXYXXPXXYTIXVLXKXXXXQLDYIXXKNPLKMSYXVGFXNKYPKRPHHRGASIPHNGV 653
P Y P Y+ VL K Q+DYI KNP KMSY VGF N YPK HHRGASIP N V
Sbjct: 463 PGWYCGPNFYSTDVLRKFAETQIDYILGKNPRKMSYVVGFGNHYPKHVHHRGASIPKNKV 522
Query: 652 KYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAANAGLVAALI 473
KY CKGG+K+RD+ KANPN +VGAMVAGPD+ DG++DVR+NYNYTEPTLA NAGLVAAL+
Sbjct: 523 KYNCKGGWKYRDSSKANPNTIVGAMVAGPDKHDGFRDVRSNYNYTEPTLAGNAGLVAALV 582
Query: 472 SISDIKTGRFGIDKNTIFSAI 410
++S + GIDKNT+FSA+
Sbjct: 583 ALSGDRD--VGIDKNTLFSAV 601
>sptrnew|CAF18445|CAF18445 Endo-1,4-beta-D-glucanase KORRIGAN (EC
3.2.1.4) (Fragment).
Length = 229
Score = 189 bits (479), Expect = 6e-47
Identities = 92/141 (65%), Positives = 104/141 (73%)
Frame = -2
Query: 832 PXXYXXPXXYTIXVLXKXXXXQLDYIXXKNPLKMSYXVGFXNKYPKRPHHRGASIPHNGV 653
P Y P Y+ L + Q+DYI KNP KMSY VGF N YPK HHRGASIP V
Sbjct: 82 PGWYCGPNFYSTDKLREFARTQIDYILGKNPRKMSYIVGFGNHYPKHVHHRGASIPKTKV 141
Query: 652 KYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAANAGLVAALI 473
KY CKGG+KWRDT K NPNILVGAMVAGPDR DG+ DVR+NYNYTEPTLA NAGLVAAL+
Sbjct: 142 KYNCKGGWKWRDTSKPNPNILVGAMVAGPDRHDGFHDVRSNYNYTEPTLAGNAGLVAALV 201
Query: 472 SISDIKTGRFGIDKNTIFSAI 410
++S K+ IDKNT+FSA+
Sbjct: 202 ALSGDKS--IPIDKNTLFSAV 220
>sptr|Q9STW8|Q9STW8 Endo-1, 4-beta-glucanase like protein.
Length = 620
Score = 171 bits (433), Expect = 1e-41
Identities = 84/141 (59%), Positives = 101/141 (71%)
Frame = -2
Query: 832 PXXYXXPXXYTIXVLXKXXXXQLDYIXXKNPLKMSYXVGFXNKYPKRPHHRGASIPHNGV 653
P Y P YT L Q+DYI KNP KMSY VG+ +YPK+ HHRGASIP N +
Sbjct: 466 PGWYCGPNFYTTEFLRNFSRSQIDYILGKNPRKMSYVVGYGQRYPKQVHHRGASIPKN-M 524
Query: 652 KYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAANAGLVAALI 473
K C GG+KW+ +KK NPN + GAMVAGPD+ DG+ D+RTNYNYTEPTLA NAGLVAAL+
Sbjct: 525 KETCTGGFKWKKSKKNNPNAINGAMVAGPDKHDGFHDIRTNYNYTEPTLAGNAGLVAALV 584
Query: 472 SISDIKTGRFGIDKNTIFSAI 410
++S K GIDKNT+FSA+
Sbjct: 585 ALSGEKAVG-GIDKNTMFSAV 604
>sptr|Q7XUK4|Q7XUK4 OSJNBa0067K08.14 protein.
Length = 623
Score = 167 bits (424), Expect = 1e-40
Identities = 79/143 (55%), Positives = 97/143 (67%), Gaps = 2/143 (1%)
Frame = -2
Query: 832 PXXYXXPXXYTIXVLXKXXXXQLDYIXXKNPLKMSYXVGFXNKYPKRPHHRGASIPHNGV 653
P Y P T+ L Q++YI NP KMSY VG+ KYP+R HHRGAS PHNG+
Sbjct: 465 PGWYCGPYFMTVDDLRSFARSQVNYILGDNPKKMSYVVGYGKKYPRRLHHRGASTPHNGI 524
Query: 652 KYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAANAGLVAALI 473
KY C GGYKWRDTK A+PN+LVGAMV GPD+ D +KD R Y EPTL NAGLVAAL+
Sbjct: 525 KYSCTGGYKWRDTKGADPNVLVGAMVGGPDKNDQFKDARLTYAQNEPTLVGNAGLVAALV 584
Query: 472 SISDIKTGR--FGIDKNTIFSAI 410
++++ G +DKNT+FSA+
Sbjct: 585 ALTNSGRGAGVTAVDKNTMFSAV 607
>sptr|O04478|O04478 F5I14.14.
Length = 623
Score = 145 bits (366), Expect = 7e-34
Identities = 71/141 (50%), Positives = 95/141 (67%)
Frame = -2
Query: 832 PXXYXXPXXYTIXVLXKXXXXQLDYIXXKNPLKMSYXVGFXNKYPKRPHHRGASIPHNGV 653
P Y P VL Q+DYI NPLKMSY VGF K+P+R HHRGA+IP++
Sbjct: 469 PGWYCGPTFVENHVLKDFAQSQIDYILGDNPLKMSYVVGFGKKFPRRVHHRGATIPNDKK 528
Query: 652 KYGCKGGYKWRDTKKANPNILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAANAGLVAALI 473
+ C+ G K+RDTK NPN + GAMV GP++ D + D+R NYN +EPTL+ NAGLVAAL+
Sbjct: 529 RRSCREGLKYRDTKNPNPNNITGAMVGGPNKFDEFHDLRNNYNASEPTLSGNAGLVAALV 588
Query: 472 SISDIKTGRFGIDKNTIFSAI 410
S++ +G IDKNT+F+++
Sbjct: 589 SLT--SSGGQQIDKNTMFNSV 607
>sptrnew|AAQ63883|AAQ63883 Cellulase.
Length = 601
Score = 121 bits (303), Expect = 1e-26
Identities = 53/115 (46%), Positives = 79/115 (68%)
Frame = -2
Query: 805 YTIXVLXKXXXXQLDYIXXKNPLKMSYXVGFXNKYPKRPHHRGASIPHNGVKYGCKGGYK 626
+++ +L Q++YI +NP+KMSY VG+ +K+P + HHR ASIP + Y C G
Sbjct: 471 FSVSMLRDFSSSQVNYILGQNPMKMSYLVGYGDKFPVQVHHRSASIPWDKRLYNCDDGKT 530
Query: 625 WRDTKKANPNILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAANAGLVAALISISD 461
W ++K NP +L+GAMV GPD D + D R+N +TEPT+++NAGLV+ALI++ D
Sbjct: 531 WLNSKNPNPQVLLGAMVGGPDTNDHFTDQRSNKRFTEPTISSNAGLVSALIALQD 585
>sptr|Q9SZ90|Q9SZ90 Cellulase-like protein.
Length = 480
Score = 109 bits (273), Expect = 4e-23
Identities = 56/120 (46%), Positives = 72/120 (60%), Gaps = 6/120 (5%)
Frame = -2
Query: 805 YTIXVLXKXXXXQLDYIXXKNPLKMSYXVGFXNKYPKRPHHRGASIP---HNGVKYGCKG 635
+T+ + Q+DYI NPLKMSY VGF KYP +PHHRG+S+P K C G
Sbjct: 361 FTVSQIRNFAKSQVDYILGNNPLKMSYMVGFGTKYPTQPHHRGSSLPSIQSKPEKIDCNG 420
Query: 634 GYKWRDTKKANPNILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAANA---GLVAALISIS 464
G+ + + NPN+ GA+V GP+ D Y D RT+Y++ EPT NA G VAALIS S
Sbjct: 421 GFSYYNFDTPNPNVHTGAIVGGPNSSDQYSDKRTDYSHAEPTTYINAAFIGSVAALISSS 480
>sptr|Q9SUS0|Q9SUS0 Putative cellulase.
Length = 479
Score = 109 bits (273), Expect = 4e-23
Identities = 54/120 (45%), Positives = 74/120 (61%), Gaps = 6/120 (5%)
Frame = -2
Query: 805 YTIXVLXKXXXXQLDYIXXKNPLKMSYXVGFXNKYPKRPHHRGASIP---HNGVKYGCKG 635
+T+ + Q+DYI NP+KMSY VGF KYP +PHHRG+S+P K C G
Sbjct: 359 FTVSQIRNFAKSQVDYILGNNPMKMSYMVGFGTKYPTQPHHRGSSLPSIQSKPEKIDCNG 418
Query: 634 GYKWRDTKKANPNILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAANA---GLVAALISIS 464
GY + ++ NPN+ +GA+V GP+ D Y D +++Y++ EPT NA G VAALIS S
Sbjct: 419 GYSYYNSDTPNPNVHIGAIVGGPNSSDQYSDKKSDYSHAEPTTYINAAFIGPVAALISSS 478
>sptr|Q93WZ1|Q93WZ1 Endo-beta-1,4-glucanase precursor (EC 3.2.1.4).
Length = 489
Score = 105 bits (262), Expect = 8e-22
Identities = 53/113 (46%), Positives = 74/113 (65%), Gaps = 4/113 (3%)
Frame = -2
Query: 802 TIXVLXKXXXXQLDYIXXKNPLKMSYXVGFXNKYPKRPHHRGASIPHNGV---KYGCKGG 632
TI +L K Q+DYI NP+KMSY VG+ YP+R HHRG+S+P + +GC GG
Sbjct: 372 TIRILAKR---QVDYILGNNPMKMSYMVGYGTNYPRRVHHRGSSLPSMAMHPQSFGCDGG 428
Query: 631 YK-WRDTKKANPNILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAANAGLVAAL 476
++ + T ANPNILVGA+V GP++ D + D RT+Y+++EP NA +V L
Sbjct: 429 FQPYYYTANANPNILVGAIVGGPNQNDFFPDERTDYSHSEPATYINAAIVGPL 481
>sptr|O82473|O82473 Endo-1,4-beta-glucanase (EC 3.2.1.4).
Length = 497
Score = 104 bits (260), Expect = 1e-21
Identities = 53/113 (46%), Positives = 70/113 (61%), Gaps = 4/113 (3%)
Frame = -2
Query: 790 LXKXXXXQLDYIXXKNPLKMSYXVGFXNKYPKRPHHRGASIP----HNGVKYGCKGGYKW 623
L + Q+DYI NP K+SY VGF +KYP R HHRG+S+P H G GC G++
Sbjct: 383 LVELAKKQVDYILGDNPAKISYMVGFGDKYPLRVHHRGSSLPSVHAHPG-HIGCNDGFQS 441
Query: 622 RDTKKANPNILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAANAGLVAALISIS 464
++ NPN+L+GA+V GPD RD ++D R NY +EP NA LV AL +S
Sbjct: 442 LNSGSPNPNVLIGAIVGGPDSRDNFEDDRNNYQQSEPATYINAPLVGALAFLS 494
>sptr|Q8LDE8|Q8LDE8 Putative beta-glucanase.
Length = 484
Score = 103 bits (258), Expect = 2e-21
Identities = 48/101 (47%), Positives = 66/101 (65%), Gaps = 3/101 (2%)
Frame = -2
Query: 769 QLDYIXXKNPLKMSYXVGFXNKYPKRPHHRGASIPHNGVK---YGCKGGYKWRDTKKANP 599
Q+DYI NP+KMSY VGF + +PKR HHR +S+P + ++ GC GG++ T+ NP
Sbjct: 375 QVDYILGDNPIKMSYMVGFSSNFPKRIHHRASSLPSHALRSQSLGCNGGFQSFYTQNPNP 434
Query: 598 NILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAANAGLVAAL 476
NIL GA+V GP++ DGY D R +Y++ EP NA V L
Sbjct: 435 NILTGAIVGGPNQNDGYPDQRDDYSHAEPATYINAAFVGPL 475
>sptr|Q9C9H5|Q9C9H5 Putative beta-glucanase, 74324-76084.
Length = 484
Score = 103 bits (258), Expect = 2e-21
Identities = 48/101 (47%), Positives = 66/101 (65%), Gaps = 3/101 (2%)
Frame = -2
Query: 769 QLDYIXXKNPLKMSYXVGFXNKYPKRPHHRGASIPHNGVK---YGCKGGYKWRDTKKANP 599
Q+DYI NP+KMSY VGF + +PKR HHR +S+P + ++ GC GG++ T+ NP
Sbjct: 375 QVDYILGDNPIKMSYMVGFSSNFPKRIHHRASSLPSHALRSQSLGCNGGFQSFYTQNPNP 434
Query: 598 NILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAANAGLVAAL 476
NIL GA+V GP++ DGY D R +Y++ EP NA V L
Sbjct: 435 NILTGAIVGGPNQNDGYPDQRDDYSHAEPATYINAAFVGPL 475
>sptr|Q43149|Q43149 Cellulase (EC 3.2.1.4) (Fragment).
Length = 508
Score = 103 bits (256), Expect = 4e-21
Identities = 54/112 (48%), Positives = 72/112 (64%), Gaps = 7/112 (6%)
Frame = -2
Query: 769 QLDYIXXKNPLKMSYXVGFXNKYPKRPHHRGASIPH---NGVKYGCKGGYK-WRDTKKAN 602
Q+DYI NP+KMSY VGF +KYP + HHRGASIP + + GC GY W + K+N
Sbjct: 381 QVDYILGNNPMKMSYMVGFGSKYPLQLHHRGASIPSMQIHPARVGCNEGYSAWYSSSKSN 440
Query: 601 PNILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAANA---GLVAALISISDIK 455
PN VGA+V GP+ D + DVR++Y++ E T NA G VAAL+ + I+
Sbjct: 441 PNTHVGAIVGGPNSNDQFNDVRSDYSHLETTTYMNAAFVGSVAALVGDNSIR 492
>sptr|Q9SML6|Q9SML6 Endo-beta-1,4-glucanase precursor (EC 3.2.1.4).
Length = 497
Score = 103 bits (256), Expect = 4e-21
Identities = 54/113 (47%), Positives = 68/113 (60%), Gaps = 4/113 (3%)
Frame = -2
Query: 790 LXKXXXXQLDYIXXKNPLKMSYXVGFXNKYPKRPHHRGASIP----HNGVKYGCKGGYKW 623
L + Q+DYI NP K+SY VGF KYP R HHRG+S+P H G GC G++
Sbjct: 380 LVELARKQVDYILGDNPAKISYMVGFGQKYPLRVHHRGSSLPSVRTHPG-HIGCNDGFQS 438
Query: 622 RDTKKANPNILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAANAGLVAALISIS 464
+ NPN+LVGA+V GPD RD ++D R NY +EP NA LV AL +S
Sbjct: 439 LYSGSPNPNVLVGAIVGGPDSRDNFEDDRNNYQQSEPATYINAPLVGALAFLS 491
>sptr|Q96546|Q96546 Endo-beta-1,4-glucanase (EC 3.2.1.4).
Length = 497
Score = 103 bits (256), Expect = 4e-21
Identities = 54/113 (47%), Positives = 68/113 (60%), Gaps = 4/113 (3%)
Frame = -2
Query: 790 LXKXXXXQLDYIXXKNPLKMSYXVGFXNKYPKRPHHRGASIP----HNGVKYGCKGGYKW 623
L + Q+DYI NP K+SY VGF KYP R HHRG+S+P H G GC G++
Sbjct: 380 LVELARKQVDYILGDNPAKISYMVGFGQKYPLRVHHRGSSLPSVRTHPG-HIGCNDGFQS 438
Query: 622 RDTKKANPNILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAANAGLVAALISIS 464
+ NPN+LVGA+V GPD RD ++D R NY +EP NA LV AL +S
Sbjct: 439 LYSGSPNPNVLVGAIVGGPDSRDNFEDDRNNYQQSEPATYINAPLVGALAFLS 491
>sptr|Q9XIY8|Q9XIY8 Endo-1,4-beta glucanase precursor.
Length = 494
Score = 102 bits (255), Expect = 5e-21
Identities = 50/105 (47%), Positives = 64/105 (60%), Gaps = 3/105 (2%)
Frame = -2
Query: 769 QLDYIXXKNPLKMSYXVGFXNKYPKRPHHRGASIPH---NGVKYGCKGGYKWRDTKKANP 599
Q+DYI NP KMSY VGF NKYP+ HHRG+S+P + + C G+++ + NP
Sbjct: 387 QVDYILGDNPAKMSYMVGFGNKYPQHVHHRGSSVPSIHAHPNRISCNDGFQYLYSSSPNP 446
Query: 598 NILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAANAGLVAALISIS 464
N+LVGA+V GPD RD + D R NY +EP NA V AL S
Sbjct: 447 NVLVGAIVGGPDNRDHFADDRNNYQQSEPATYINAPFVGALAFFS 491
>sw|P22503|GUN_PHAVU Endoglucanase precursor (EC 3.2.1.4)
(Endo-1,4-beta-glucanase) (Abscission cellulase).
Length = 496
Score = 102 bits (253), Expect = 9e-21
Identities = 50/117 (42%), Positives = 72/117 (61%), Gaps = 4/117 (3%)
Frame = -2
Query: 805 YTIXVLXKXXXXQLDYIXXKNPLKMSYXVGFXNKYPKRPHHRGASIPH---NGVKYGCKG 635
+T + Q++YI KNP+KMSY VGF +KYPK+ HHRG+SIP + K GC
Sbjct: 376 FTASQIRGFAKTQVEYILGKNPMKMSYMVGFGSKYPKQLHHRGSSIPSIKVHPAKVGCNA 435
Query: 634 GYK-WRDTKKANPNILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAANAGLVAALISI 467
G + ++ NPN VGA+V GPD D + D R++Y++ EPT NA VA++ ++
Sbjct: 436 GLSDYYNSANPNPNTHVGAIVGGPDSNDRFNDARSDYSHAEPTTYINAAFVASISAL 492
>sptr|O23134|O23134 Beta-glucanase.
Length = 484
Score = 102 bits (253), Expect = 9e-21
Identities = 48/101 (47%), Positives = 64/101 (63%), Gaps = 3/101 (2%)
Frame = -2
Query: 769 QLDYIXXKNPLKMSYXVGFXNKYPKRPHHRGASIPHNGVK---YGCKGGYKWRDTKKANP 599
Q+DY+ NP+KMSY VGF + +PKR HHRG+S+P V+ GC GG++ T+ NP
Sbjct: 375 QVDYVLGVNPMKMSYMVGFSSNFPKRIHHRGSSLPSRAVRSNSLGCNGGFQSFRTQNPNP 434
Query: 598 NILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAANAGLVAAL 476
NIL GA+V GP++ D Y D R +Y +EP NA V L
Sbjct: 435 NILTGAIVGGPNQNDEYPDQRDDYTRSEPATYINAAFVGPL 475
>sptr|Q40763|Q40763 Cellulase precursor.
Length = 494
Score = 101 bits (251), Expect = 2e-20
Identities = 47/105 (44%), Positives = 63/105 (60%), Gaps = 3/105 (2%)
Frame = -2
Query: 769 QLDYIXXKNPLKMSYXVGFXNKYPKRPHHRGASIPH---NGVKYGCKGGYKWRDTKKANP 599
Q+DYI NP +MSY VGF N+YP+ HHRG+S+P + C G+++ + NP
Sbjct: 387 QVDYILGDNPARMSYMVGFGNRYPQHVHHRGSSVPSIHDTRIGISCNDGFQFLYSSSPNP 446
Query: 598 NILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAANAGLVAALISIS 464
N+LVGA++ GPD RD + D R NY +EP NA V AL S
Sbjct: 447 NVLVGAIIGGPDNRDNFADSRNNYQQSEPATYINAPFVGALAFFS 491
>sw|P05522|GUN1_PERAE Endoglucanase 1 precursor (EC 3.2.1.4)
(Endo-1,4-beta-glucanase) (Abscission cellulase 1).
Length = 494
Score = 100 bits (250), Expect = 2e-20
Identities = 49/101 (48%), Positives = 63/101 (62%), Gaps = 3/101 (2%)
Frame = -2
Query: 769 QLDYIXXKNPLKMSYXVGFXNKYPKRPHHRGASIPHNGV---KYGCKGGYKWRDTKKANP 599
Q+DYI +NP KMSY VGF +YP+ HHRG+S+P V C G+++ + NP
Sbjct: 384 QVDYILGQNPAKMSYMVGFGERYPQHVHHRGSSLPSVQVHPNSIPCNAGFQYLYSSPPNP 443
Query: 598 NILVGAMVAGPDRRDGYKDVRTNYNYTEPTLAANAGLVAAL 476
NILVGA++ GPD RD + D R NY +EP NA LV AL
Sbjct: 444 NILVGAILGGPDNRDSFSDDRNNYQQSEPATYINAPLVGAL 484
Database: /db/uniprot/tmp/swall
Posted date: Mar 5, 2004 7:45 PM
Number of letters in database: 442,889,342
Number of sequences in database: 1,395,590
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 566,034,691
Number of Sequences: 1395590
Number of extensions: 10931450
Number of successful extensions: 25436
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 24428
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25273
length of database: 442,889,342
effective HSP length: 122
effective length of database: 272,627,362
effective search space used: 43347750558
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

