BLASTX 2.2.6 [Apr-09-2003]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
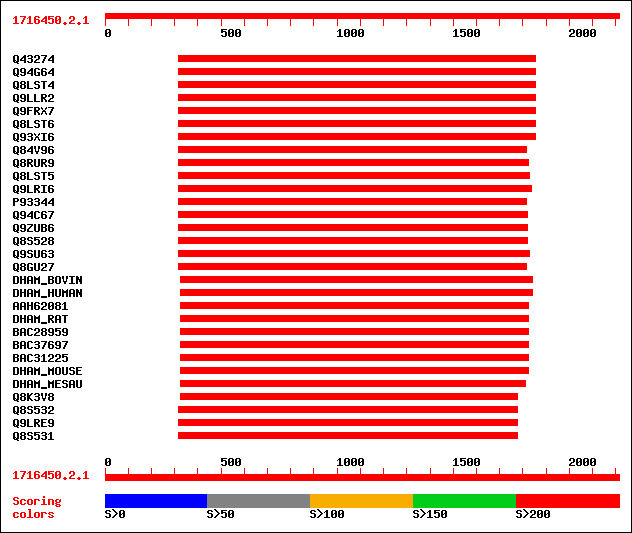
Query= 1716450.2.1
(2214 letters)
Database: /db/uniprot/tmp/swall
1,395,590 sequences; 442,889,342 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sptr|Q43274|Q43274 T CYTOPLASM MALE sterility RESTORER factor 2 ... 1031 0.0
sptr|Q94G64|Q94G64 T-cytoplasm male sterility restorer factor 2. 1025 0.0
sptr|Q8LST4|Q8LST4 Mitochondrial aldehyde dehydrogenase. 1003 0.0
sptr|Q9LLR2|Q9LLR2 Aldehyde dehydrogenase. 969 0.0
sptr|Q9FRX7|Q9FRX7 Aldehyde dehydrogenase ALDH2b. 967 0.0
sptr|Q8LST6|Q8LST6 Mitochondrial aldehyde dehydrogenase. 949 0.0
sptr|Q93XI6|Q93XI6 Mitochondrial aldehyde dehydrogenase ALDH2. 946 0.0
sptr|Q84V96|Q84V96 Aldehyde dehydrogenase 1 precursor (EC 1.2.1.3). 861 0.0
sptr|Q8RUR9|Q8RUR9 Mitochondrial aldehyde dehydrogenase RF2B. 860 0.0
sptr|Q8LST5|Q8LST5 Mitochondrial aldehyde dehydrogenase. 860 0.0
sptr|Q9LRI6|Q9LRI6 Mitochondrial aldehyde dehydrogenase ALDH2a. 859 0.0
sptr|P93344|P93344 Aldehyde dehydrogenase (NAD+) (EC 1.2.1.3). 857 0.0
sptr|Q94C67|Q94C67 Putative aldehyde dehydrogenase NAD+. 855 0.0
sptr|Q9ZUB6|Q9ZUB6 F5O8.35 protein. 847 0.0
sptr|Q8S528|Q8S528 Mitochondrial aldehyde dehydrogenase. 837 0.0
sptr|Q9SU63|Q9SU63 Aldehyde dehydrogenase (NAD+)-like protein (E... 833 0.0
sptr|Q8GU27|Q8GU27 Aldehyde dehydrogenase. 612 e-173
sw|P20000|DHAM_BOVIN Aldehyde dehydrogenase, mitochondrial precu... 611 e-173
sw|P05091|DHAM_HUMAN Aldehyde dehydrogenase, mitochondrial precu... 610 e-173
sptrnew|AAH62081|AAH62081 Aldehyde dehydrogenase 2. 607 e-172
sw|P11884|DHAM_RAT Aldehyde dehydrogenase, mitochondrial precurs... 607 e-172
sptrnew|BAC28959|BAC28959 12 days embryo embryonic body between ... 607 e-172
sptrnew|BAC37697|BAC37697 Adult male spinal cord cDNA, RIKEN ful... 607 e-172
sptrnew|BAC31225|BAC31225 3 days neonate thymus cDNA, RIKEN full... 607 e-172
sw|P47738|DHAM_MOUSE Aldehyde dehydrogenase, mitochondrial precu... 607 e-172
sw|P81178|DHAM_MESAU Aldehyde dehydrogenase, mitochondrial (EC 1... 606 e-172
sptr|Q8K3V8|Q8K3V8 Mitochondrial aldehyde dehydrogenase (Fragment). 605 e-172
sptr|Q8S532|Q8S532 Cytosolic aldehyde dehydrogenase RF2C. 605 e-171
sptr|Q9LRE9|Q9LRE9 Cytosolic aldehyde dehydrogenase. 601 e-170
sptr|Q8S531|Q8S531 Cytosolic aldehyde dehydrogenase RF2C. 600 e-170
>sptr|Q43274|Q43274 T CYTOPLASM MALE sterility RESTORER factor 2 (EC
1.2.1.3) (Aldehyde dehydrogenase (NAD+)).
Length = 549
Score = 1031 bits (2666), Expect = 0.0
Identities = 512/513 (99%), Positives = 513/513 (100%)
Frame = -2
Query: 1853 DGMHRLLPGVLQRFSTAAAVEEPITPSVHVNYTKLLINGNFVDSASGKTFPTLDPRTGEV 1674
DGMHRLLPGVLQRFSTAAAVEEPITPSVHVNYTKLLINGNFVDSASGKTFPTLDPRTGEV
Sbjct: 37 DGMHRLLPGVLQRFSTAAAVEEPITPSVHVNYTKLLINGNFVDSASGKTFPTLDPRTGEV 96
Query: 1673 IAHVAEGDAEDINRAVAAARKAFDEGPWPKMTAYERSRILLRFADLIEKHNDELAALETW 1494
IAHVAEGDAEDINRAVAAARKAFDEGPWPKMTAYERSRILLRFADLIEKHNDELAALETW
Sbjct: 97 IAHVAEGDAEDINRAVAAARKAFDEGPWPKMTAYERSRILLRFADLIEKHNDELAALETW 156
Query: 1493 DNGKPYEQAAQIEVPMVARLMRYYAGWADKIHGLIVPADGPHHVQILHEPIGVAGQIIPW 1314
DNGKPYEQAAQIEVPMVARLMRYYAGWADKIHGLIVPADGPHHVQILHEPIGVAGQIIPW
Sbjct: 157 DNGKPYEQAAQIEVPMVARLMRYYAGWADKIHGLIVPADGPHHVQILHEPIGVAGQIIPW 216
Query: 1313 NFPLLMFAWKVGPALACGNTLVLKTAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTA 1134
NFPLLM+AWKVGPALACGNTLVLKTAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTA
Sbjct: 217 NFPLLMYAWKVGPALACGNTLVLKTAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTA 276
Query: 1133 GAALASHMDVDKIAFTGSTDTGKIILELAAKSNLKTVTLELGGKSPFIIMDDADVDHAVE 954
GAALASHMDVDKIAFTGSTDTGKIILELAAKSNLKTVTLELGGKSPFIIMDDADVDHAVE
Sbjct: 277 GAALASHMDVDKIAFTGSTDTGKIILELAAKSNLKTVTLELGGKSPFIIMDDADVDHAVE 336
Query: 953 LAHFALFFNQGQCCCAGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDE 774
LAHFALFFNQGQCCCAGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDE
Sbjct: 337 LAHFALFFNQGQCCCAGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDE 396
Query: 773 QFNKILRYIRYGVDGGATLVTGGDRLGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSIL 594
QFNKILRYIRYGVDGGATLVTGGDRLGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSIL
Sbjct: 397 QFNKILRYIRYGVDGGATLVTGGDRLGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSIL 456
Query: 593 KFKDLNEVIKRANASQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGY 414
KFKDLNEVIKRANASQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGY
Sbjct: 457 KFKDLNEVIKRANASQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGY 516
Query: 413 KMSGIGREKGVDSLKNYLQVKAVVTPIKNAAWL 315
KMSGIGREKGVDSLKNYLQVKAVVTPIKNAAWL
Sbjct: 517 KMSGIGREKGVDSLKNYLQVKAVVTPIKNAAWL 549
>sptr|Q94G64|Q94G64 T-cytoplasm male sterility restorer factor 2.
Length = 549
Score = 1025 bits (2649), Expect = 0.0
Identities = 509/513 (99%), Positives = 511/513 (99%)
Frame = -2
Query: 1853 DGMHRLLPGVLQRFSTAAAVEEPITPSVHVNYTKLLINGNFVDSASGKTFPTLDPRTGEV 1674
DGMHRLLPGVLQRFSTAAAVEEPITPSVHVNYTKLLINGNFVDSASGKTFPTLDPRTGEV
Sbjct: 37 DGMHRLLPGVLQRFSTAAAVEEPITPSVHVNYTKLLINGNFVDSASGKTFPTLDPRTGEV 96
Query: 1673 IAHVAEGDAEDINRAVAAARKAFDEGPWPKMTAYERSRILLRFADLIEKHNDELAALETW 1494
IAHVAEGDAEDINRAVAAARKAFDEGPWPKMTAYERSRILLRFADLIEKHNDELAALETW
Sbjct: 97 IAHVAEGDAEDINRAVAAARKAFDEGPWPKMTAYERSRILLRFADLIEKHNDELAALETW 156
Query: 1493 DNGKPYEQAAQIEVPMVARLMRYYAGWADKIHGLIVPADGPHHVQILHEPIGVAGQIIPW 1314
DNGKPYEQAAQIEVPMVARLMRYYAGWADKIHGLIVPADGPHHVQILHEPIGVAGQIIPW
Sbjct: 157 DNGKPYEQAAQIEVPMVARLMRYYAGWADKIHGLIVPADGPHHVQILHEPIGVAGQIIPW 216
Query: 1313 NFPLLMFAWKVGPALACGNTLVLKTAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTA 1134
NFPLLM+AWKVGPALACGNTLVLKTAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTA
Sbjct: 217 NFPLLMYAWKVGPALACGNTLVLKTAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTA 276
Query: 1133 GAALASHMDVDKIAFTGSTDTGKIILELAAKSNLKTVTLELGGKSPFIIMDDADVDHAVE 954
GAALASHMDVDKIAFTGSTDTGKIILELAAKSNLKTVTLELGGKS FIIMDDADVDHAVE
Sbjct: 277 GAALASHMDVDKIAFTGSTDTGKIILELAAKSNLKTVTLELGGKSSFIIMDDADVDHAVE 336
Query: 953 LAHFALFFNQGQCCCAGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDE 774
LAHFALFFNQGQCCCAGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDE
Sbjct: 337 LAHFALFFNQGQCCCAGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDE 396
Query: 773 QFNKILRYIRYGVDGGATLVTGGDRLGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSIL 594
QF KILRYIRYGVDGGATLVTGGDRLGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSIL
Sbjct: 397 QFKKILRYIRYGVDGGATLVTGGDRLGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSIL 456
Query: 593 KFKDLNEVIKRANASQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGY 414
KFKDLNEVIKRANASQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGY
Sbjct: 457 KFKDLNEVIKRANASQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGY 516
Query: 413 KMSGIGREKGVDSLKNYLQVKAVVTPIKNAAWL 315
KMSG+GREKGVDSLKNYLQVKAVVTPIKNAAWL
Sbjct: 517 KMSGMGREKGVDSLKNYLQVKAVVTPIKNAAWL 549
>sptr|Q8LST4|Q8LST4 Mitochondrial aldehyde dehydrogenase.
Length = 547
Score = 1003 bits (2593), Expect = 0.0
Identities = 493/513 (96%), Positives = 505/513 (98%)
Frame = -2
Query: 1853 DGMHRLLPGVLQRFSTAAAVEEPITPSVHVNYTKLLINGNFVDSASGKTFPTLDPRTGEV 1674
DGM LLPGVLQRFSTAAAVEEPITPSV VNYTKLLINGNFVD+ASGKTFPTLDPRTGEV
Sbjct: 35 DGMRGLLPGVLQRFSTAAAVEEPITPSVQVNYTKLLINGNFVDAASGKTFPTLDPRTGEV 94
Query: 1673 IAHVAEGDAEDINRAVAAARKAFDEGPWPKMTAYERSRILLRFADLIEKHNDELAALETW 1494
IAHVAEGDAEDINRAVAAARKAFDEGPWPKMTAYERSRILLRFADLIEKHNDELAALETW
Sbjct: 95 IAHVAEGDAEDINRAVAAARKAFDEGPWPKMTAYERSRILLRFADLIEKHNDELAALETW 154
Query: 1493 DNGKPYEQAAQIEVPMVARLMRYYAGWADKIHGLIVPADGPHHVQILHEPIGVAGQIIPW 1314
DNGKPYEQAA IEVPMVARLMRYYAGWADKIHGL+VPADGPHHVQILHEPIGVAGQIIPW
Sbjct: 155 DNGKPYEQAAHIEVPMVARLMRYYAGWADKIHGLVVPADGPHHVQILHEPIGVAGQIIPW 214
Query: 1313 NFPLLMFAWKVGPALACGNTLVLKTAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTA 1134
NFPLLMFAWKVGPALACGNTLVLKTAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTA
Sbjct: 215 NFPLLMFAWKVGPALACGNTLVLKTAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTA 274
Query: 1133 GAALASHMDVDKIAFTGSTDTGKIILELAAKSNLKTVTLELGGKSPFIIMDDADVDHAVE 954
GAALASHMDVDK+AFTGSTDTGK++LELAA+SNLKTVTLELGGKSPFIIMDDAD+DHAVE
Sbjct: 275 GAALASHMDVDKLAFTGSTDTGKVVLELAARSNLKTVTLELGGKSPFIIMDDADIDHAVE 334
Query: 953 LAHFALFFNQGQCCCAGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDE 774
LAHFALFFNQGQCCCAGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDE
Sbjct: 335 LAHFALFFNQGQCCCAGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDE 394
Query: 773 QFNKILRYIRYGVDGGATLVTGGDRLGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSIL 594
QFNKILRYIR GVD GA LVTGGDRLG+KG+YIQPTIFSDVQDGMKIAQEEIFGPVQSIL
Sbjct: 395 QFNKILRYIRSGVDSGANLVTGGDRLGEKGYYIQPTIFSDVQDGMKIAQEEIFGPVQSIL 454
Query: 593 KFKDLNEVIKRANASQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGY 414
KFKDLNEVIKRANAS YGLAAGVFTNSLDTANTLTRALRAGTVW+NCFD+FDAAIPFGGY
Sbjct: 455 KFKDLNEVIKRANASPYGLAAGVFTNSLDTANTLTRALRAGTVWINCFDIFDAAIPFGGY 514
Query: 413 KMSGIGREKGVDSLKNYLQVKAVVTPIKNAAWL 315
KMSGIGREKG+DSLKNYLQVKAVVTPIKNAAWL
Sbjct: 515 KMSGIGREKGIDSLKNYLQVKAVVTPIKNAAWL 547
>sptr|Q9LLR2|Q9LLR2 Aldehyde dehydrogenase.
Length = 549
Score = 969 bits (2506), Expect = 0.0
Identities = 472/513 (92%), Positives = 495/513 (96%)
Frame = -2
Query: 1853 DGMHRLLPGVLQRFSTAAAVEEPITPSVHVNYTKLLINGNFVDSASGKTFPTLDPRTGEV 1674
DG LLPG+LQRFSTAA EEPI+P V VNYT+LLI+G FVDSASGKTFPTLDPRTGE+
Sbjct: 37 DGTQGLLPGILQRFSTAAVAEEPISPPVQVNYTQLLIDGKFVDSASGKTFPTLDPRTGEL 96
Query: 1673 IAHVAEGDAEDINRAVAAARKAFDEGPWPKMTAYERSRILLRFADLIEKHNDELAALETW 1494
IAHVAEGDAEDINRAV AARKAFDEGPWPKMTAYERSRILLRFADLIEKHNDE+AALETW
Sbjct: 97 IAHVAEGDAEDINRAVHAARKAFDEGPWPKMTAYERSRILLRFADLIEKHNDEIAALETW 156
Query: 1493 DNGKPYEQAAQIEVPMVARLMRYYAGWADKIHGLIVPADGPHHVQILHEPIGVAGQIIPW 1314
DNGKPY QAAQIEVPMVARLMRYYAGWADKIHGL+VPADGPHHVQ+LHEPIGVAGQIIPW
Sbjct: 157 DNGKPYAQAAQIEVPMVARLMRYYAGWADKIHGLVVPADGPHHVQVLHEPIGVAGQIIPW 216
Query: 1313 NFPLLMFAWKVGPALACGNTLVLKTAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTA 1134
NFPLLMFAWKVGPALACGNT+VLKTAEQTPLSAL+ SKLLHEAGLP+GVVNVVSGFGPTA
Sbjct: 217 NFPLLMFAWKVGPALACGNTVVLKTAEQTPLSALFASKLLHEAGLPDGVVNVVSGFGPTA 276
Query: 1133 GAALASHMDVDKIAFTGSTDTGKIILELAAKSNLKTVTLELGGKSPFIIMDDADVDHAVE 954
GAALASHMDVDKIAFTGSTDTGK++LELAA+SNLK+VTLELGGKSPFIIMDDADVDHAVE
Sbjct: 277 GAALASHMDVDKIAFTGSTDTGKVVLELAARSNLKSVTLELGGKSPFIIMDDADVDHAVE 336
Query: 953 LAHFALFFNQGQCCCAGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDE 774
LAHFALFFNQGQCCCAGSRTFVHER+YDEFVEKAKARALKRVVGDPF+ GVEQGPQIDDE
Sbjct: 337 LAHFALFFNQGQCCCAGSRTFVHERIYDEFVEKAKARALKRVVGDPFKNGVEQGPQIDDE 396
Query: 773 QFNKILRYIRYGVDGGATLVTGGDRLGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSIL 594
QFNKILRYI+YGVD GA LVTGGDRLGDKG+YIQPTIFSDVQD M+IAQEEIFGPVQSIL
Sbjct: 397 QFNKILRYIKYGVDSGANLVTGGDRLGDKGYYIQPTIFSDVQDNMRIAQEEIFGPVQSIL 456
Query: 593 KFKDLNEVIKRANASQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGY 414
KF DLNEVIKRANASQYGLAAGVFTN+L+TANTLTRALR GTVWVNCFDVFDAAIPFGGY
Sbjct: 457 KFNDLNEVIKRANASQYGLAAGVFTNNLNTANTLTRALRVGTVWVNCFDVFDAAIPFGGY 516
Query: 413 KMSGIGREKGVDSLKNYLQVKAVVTPIKNAAWL 315
K SGIGREKG+DSLKNYLQVKAVVTPIKNAAWL
Sbjct: 517 KQSGIGREKGIDSLKNYLQVKAVVTPIKNAAWL 549
>sptr|Q9FRX7|Q9FRX7 Aldehyde dehydrogenase ALDH2b.
Length = 549
Score = 967 bits (2501), Expect = 0.0
Identities = 471/513 (91%), Positives = 494/513 (96%)
Frame = -2
Query: 1853 DGMHRLLPGVLQRFSTAAAVEEPITPSVHVNYTKLLINGNFVDSASGKTFPTLDPRTGEV 1674
DG LLPG+LQRFSTAA EEPI+P V VNYT+LLI+G FVDSASGKTFPTLDPRTGE+
Sbjct: 37 DGTQGLLPGILQRFSTAAVAEEPISPPVQVNYTQLLIDGKFVDSASGKTFPTLDPRTGEL 96
Query: 1673 IAHVAEGDAEDINRAVAAARKAFDEGPWPKMTAYERSRILLRFADLIEKHNDELAALETW 1494
IAHVAEGDAEDINRAV AARKAFDEGPWPKMTAYERSRILLRFADLIEKHNDE+AALETW
Sbjct: 97 IAHVAEGDAEDINRAVHAARKAFDEGPWPKMTAYERSRILLRFADLIEKHNDEIAALETW 156
Query: 1493 DNGKPYEQAAQIEVPMVARLMRYYAGWADKIHGLIVPADGPHHVQILHEPIGVAGQIIPW 1314
DNGKPY QAA IEVPMVARLMRYYAGWADKIHGL+VPADGPHHVQ+LHEPIGVAGQIIPW
Sbjct: 157 DNGKPYAQAANIEVPMVARLMRYYAGWADKIHGLVVPADGPHHVQVLHEPIGVAGQIIPW 216
Query: 1313 NFPLLMFAWKVGPALACGNTLVLKTAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTA 1134
NFPLLMFAWKVGPALACGNT+VLKTAEQTPLSAL+ SKLLHEAGLP+GVVNVVSGFGPTA
Sbjct: 217 NFPLLMFAWKVGPALACGNTVVLKTAEQTPLSALFASKLLHEAGLPDGVVNVVSGFGPTA 276
Query: 1133 GAALASHMDVDKIAFTGSTDTGKIILELAAKSNLKTVTLELGGKSPFIIMDDADVDHAVE 954
GAALASHMDVDKIAFTGSTDTGK++LELAA+SNLK+VTLELGGKSPFIIMDDADVDHAVE
Sbjct: 277 GAALASHMDVDKIAFTGSTDTGKVVLELAARSNLKSVTLELGGKSPFIIMDDADVDHAVE 336
Query: 953 LAHFALFFNQGQCCCAGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDE 774
LAHFALFFNQGQCCCAGSRTFVHER+YDEFVEKAKARALKRVVGDPF+ GVEQGPQIDDE
Sbjct: 337 LAHFALFFNQGQCCCAGSRTFVHERIYDEFVEKAKARALKRVVGDPFKNGVEQGPQIDDE 396
Query: 773 QFNKILRYIRYGVDGGATLVTGGDRLGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSIL 594
QFNKILRYI+YGVD GA LVTGGDRLGDKG+YIQPTIFSDVQD M+IAQEEIFGPVQSIL
Sbjct: 397 QFNKILRYIKYGVDSGANLVTGGDRLGDKGYYIQPTIFSDVQDNMRIAQEEIFGPVQSIL 456
Query: 593 KFKDLNEVIKRANASQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGY 414
KF DLNEVIKRANASQYGLAAGVFTN+L+TANTLTRALR GTVWVNCFDVFDAAIPFGGY
Sbjct: 457 KFNDLNEVIKRANASQYGLAAGVFTNNLNTANTLTRALRVGTVWVNCFDVFDAAIPFGGY 516
Query: 413 KMSGIGREKGVDSLKNYLQVKAVVTPIKNAAWL 315
K SGIGREKG+DSLKNYLQVKAVVTPIKNAAWL
Sbjct: 517 KQSGIGREKGIDSLKNYLQVKAVVTPIKNAAWL 549
>sptr|Q8LST6|Q8LST6 Mitochondrial aldehyde dehydrogenase.
Length = 549
Score = 949 bits (2454), Expect = 0.0
Identities = 459/513 (89%), Positives = 490/513 (95%)
Frame = -2
Query: 1853 DGMHRLLPGVLQRFSTAAAVEEPITPSVHVNYTKLLINGNFVDSASGKTFPTLDPRTGEV 1674
DG LLPG+LQRF TAAA EEPI+PSV V+ T+LLING FVD+ASGKTFPTLDPRTGEV
Sbjct: 37 DGARGLLPGLLQRFGTAAAAEEPISPSVQVSETQLLINGKFVDAASGKTFPTLDPRTGEV 96
Query: 1673 IAHVAEGDAEDINRAVAAARKAFDEGPWPKMTAYERSRILLRFADLIEKHNDELAALETW 1494
IA V+EGDAED++RAV AARKAFD GPWPKMTAYERSRILLRFADLIEKHND++AALETW
Sbjct: 97 IARVSEGDAEDVDRAVIAARKAFDHGPWPKMTAYERSRILLRFADLIEKHNDDIAALETW 156
Query: 1493 DNGKPYEQAAQIEVPMVARLMRYYAGWADKIHGLIVPADGPHHVQILHEPIGVAGQIIPW 1314
DNGKPYEQAAQIEVPM+ARLMRYYAGW DKIHGLIVPADGPHHVQ+LHEPIGV GQIIPW
Sbjct: 157 DNGKPYEQAAQIEVPMLARLMRYYAGWTDKIHGLIVPADGPHHVQVLHEPIGVVGQIIPW 216
Query: 1313 NFPLLMFAWKVGPALACGNTLVLKTAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTA 1134
NFPLLM+ WKVGPALACGNT+VLKTAEQTPLSALY+SKLLHEAGLPEGV+N+VSGFGPTA
Sbjct: 217 NFPLLMYGWKVGPALACGNTIVLKTAEQTPLSALYVSKLLHEAGLPEGVLNIVSGFGPTA 276
Query: 1133 GAALASHMDVDKIAFTGSTDTGKIILELAAKSNLKTVTLELGGKSPFIIMDDADVDHAVE 954
GAALASHMDVDKIAFTGSTDTGK+ILEL+A+SNLK VTLELGGKSPFI+MDDAD+D AVE
Sbjct: 277 GAALASHMDVDKIAFTGSTDTGKVILELSARSNLKPVTLELGGKSPFIVMDDADIDQAVE 336
Query: 953 LAHFALFFNQGQCCCAGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDE 774
LAHFALFFNQGQCCCAGSRTFVHERVYDEFVEK+KARALKRVVGDPFRKGVEQGPQIDDE
Sbjct: 337 LAHFALFFNQGQCCCAGSRTFVHERVYDEFVEKSKARALKRVVGDPFRKGVEQGPQIDDE 396
Query: 773 QFNKILRYIRYGVDGGATLVTGGDRLGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSIL 594
QF KILRYI+ GVD GATLVTGGD+LGDKG+YIQPTIFSDVQDGMKIAQEEIFGPVQSI
Sbjct: 397 QFKKILRYIKSGVDSGATLVTGGDKLGDKGYYIQPTIFSDVQDGMKIAQEEIFGPVQSIF 456
Query: 593 KFKDLNEVIKRANASQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGY 414
KF DLNEVIKRANASQYGLAAGVFTN+LDTANTLTRALRAGT+WVNCFD+FDAAIPFGGY
Sbjct: 457 KFNDLNEVIKRANASQYGLAAGVFTNNLDTANTLTRALRAGTIWVNCFDIFDAAIPFGGY 516
Query: 413 KMSGIGREKGVDSLKNYLQVKAVVTPIKNAAWL 315
KMSGIGREKG+DSLKNYLQVKAVVT +KN AWL
Sbjct: 517 KMSGIGREKGIDSLKNYLQVKAVVTALKNPAWL 549
>sptr|Q93XI6|Q93XI6 Mitochondrial aldehyde dehydrogenase ALDH2.
Length = 549
Score = 946 bits (2446), Expect = 0.0
Identities = 457/513 (89%), Positives = 487/513 (94%)
Frame = -2
Query: 1853 DGMHRLLPGVLQRFSTAAAVEEPITPSVHVNYTKLLINGNFVDSASGKTFPTLDPRTGEV 1674
DG LLPG+LQRF TAAA EEPI+PSV V T+LLING FVD+ASGKTFPTLDPRTGEV
Sbjct: 37 DGARGLLPGLLQRFGTAAAAEEPISPSVQVGETQLLINGKFVDAASGKTFPTLDPRTGEV 96
Query: 1673 IAHVAEGDAEDINRAVAAARKAFDEGPWPKMTAYERSRILLRFADLIEKHNDELAALETW 1494
IA V+EGDAED++RAV AARKAFDEGPWPKMTAYERSRILLRFADLIEKHNDE+AALETW
Sbjct: 97 IARVSEGDAEDVDRAVVAARKAFDEGPWPKMTAYERSRILLRFADLIEKHNDEIAALETW 156
Query: 1493 DNGKPYEQAAQIEVPMVARLMRYYAGWADKIHGLIVPADGPHHVQILHEPIGVAGQIIPW 1314
DNGKPYEQAA IEVPM+ARLMRYYAGW DKIHGLIVPADGPHHVQ+LHEPIGV GQIIPW
Sbjct: 157 DNGKPYEQAAHIEVPMLARLMRYYAGWTDKIHGLIVPADGPHHVQVLHEPIGVVGQIIPW 216
Query: 1313 NFPLLMFAWKVGPALACGNTLVLKTAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTA 1134
NFPLLM+ WKVGPALACGNT+VLKTAEQTPLSALY+SKLLHEAGLPEGV+N++SGFGPTA
Sbjct: 217 NFPLLMYGWKVGPALACGNTIVLKTAEQTPLSALYVSKLLHEAGLPEGVLNIISGFGPTA 276
Query: 1133 GAALASHMDVDKIAFTGSTDTGKIILELAAKSNLKTVTLELGGKSPFIIMDDADVDHAVE 954
GAALA HMDVDKIAFTGSTDTGK+ILEL+A+SNLK VTLELGGKSPFI+MDDAD+D AVE
Sbjct: 277 GAALAGHMDVDKIAFTGSTDTGKVILELSARSNLKAVTLELGGKSPFIVMDDADIDQAVE 336
Query: 953 LAHFALFFNQGQCCCAGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDE 774
LAHFALFFNQGQCCCAGSRTFVHERVYDEFVEK+KARALKRVVGDPFRKGVEQGPQIDDE
Sbjct: 337 LAHFALFFNQGQCCCAGSRTFVHERVYDEFVEKSKARALKRVVGDPFRKGVEQGPQIDDE 396
Query: 773 QFNKILRYIRYGVDGGATLVTGGDRLGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSIL 594
QF KILRYI+ GVD GATLVTGGD+LGDKG+YIQPTIFSDVQD MKIAQEEIFGPVQSI
Sbjct: 397 QFKKILRYIKSGVDSGATLVTGGDKLGDKGYYIQPTIFSDVQDDMKIAQEEIFGPVQSIF 456
Query: 593 KFKDLNEVIKRANASQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGY 414
KF DLNEVIKRANASQYGLAAGVFTN+LDTANTLTRALRAGT+WVNCFD+FDAAIPFGGY
Sbjct: 457 KFNDLNEVIKRANASQYGLAAGVFTNNLDTANTLTRALRAGTIWVNCFDIFDAAIPFGGY 516
Query: 413 KMSGIGREKGVDSLKNYLQVKAVVTPIKNAAWL 315
KMSGIGREKG+DSLKNYLQVKAVVT +KN AWL
Sbjct: 517 KMSGIGREKGIDSLKNYLQVKAVVTALKNPAWL 549
>sptr|Q84V96|Q84V96 Aldehyde dehydrogenase 1 precursor (EC 1.2.1.3).
Length = 542
Score = 861 bits (2224), Expect = 0.0
Identities = 413/501 (82%), Positives = 460/501 (91%)
Frame = -2
Query: 1817 RFSTAAAVEEPITPSVHVNYTKLLINGNFVDSASGKTFPTLDPRTGEVIAHVAEGDAEDI 1638
R+STAA VEE ITP V +NYT+ LING FVD+ASGKTFP DPRTG+VIAHVAEGDAED+
Sbjct: 42 RYSTAAVVEELITPQVSINYTQHLINGKFVDAASGKTFPAYDPRTGDVIAHVAEGDAEDV 101
Query: 1637 NRAVAAARKAFDEGPWPKMTAYERSRILLRFADLIEKHNDELAALETWDNGKPYEQAAQI 1458
NRAVAAARKAFDEGPWP+MTAYERSRILLRFADL+EKHNDE+AALETW+NGKPYEQAA+
Sbjct: 102 NRAVAAARKAFDEGPWPRMTAYERSRILLRFADLVEKHNDEIAALETWNNGKPYEQAAKA 161
Query: 1457 EVPMVARLMRYYAGWADKIHGLIVPADGPHHVQILHEPIGVAGQIIPWNFPLLMFAWKVG 1278
EVP RL RYYAGWADKIHGL VPADG +HVQ LHEPIGVAGQIIPWNFPLLMFAWKVG
Sbjct: 162 EVPTFVRLFRYYAGWADKIHGLTVPADGNYHVQTLHEPIGVAGQIIPWNFPLLMFAWKVG 221
Query: 1277 PALACGNTLVLKTAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTAGAALASHMDVDK 1098
PALACGNT+VLKTAEQTPL+AL ++KLLHEAGLP GV+N+VSG+GPTAGA LASHMDVDK
Sbjct: 222 PALACGNTIVLKTAEQTPLTALVVAKLLHEAGLPPGVLNIVSGYGPTAGAPLASHMDVDK 281
Query: 1097 IAFTGSTDTGKIILELAAKSNLKTVTLELGGKSPFIIMDDADVDHAVELAHFALFFNQGQ 918
+AFTGSTDTGK++L LAAKSNLK VTLELGGKSPFI+ +DADVD AVELAHFALFFNQGQ
Sbjct: 282 LAFTGSTDTGKVVLGLAAKSNLKPVTLELGGKSPFIVCEDADVDKAVELAHFALFFNQGQ 341
Query: 917 CCCAGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDEQFNKILRYIRYG 738
CCCAGSRTFVHERVYDEF+EK+K RAL+RVVGDPF+KGVEQGPQID EQF KILRYI+ G
Sbjct: 342 CCCAGSRTFVHERVYDEFLEKSKKRALRRVVGDPFKKGVEQGPQIDTEQFEKILRYIKSG 401
Query: 737 VDGGATLVTGGDRLGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSILKFKDLNEVIKRA 558
++ ATL GGDRLG KGFY+QPT+FS+VQD M IAQ+EIFGPVQ+I KFK+++EVI+RA
Sbjct: 402 IESNATLECGGDRLGSKGFYVQPTVFSNVQDDMLIAQDEIFGPVQTIFKFKEIDEVIRRA 461
Query: 557 NASQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGYKMSGIGREKGVD 378
N+++YGLAAGVFT +L TANTL RALRAGTVW+NCFDVFDAAIPFGGYKMSGIGREKG+
Sbjct: 462 NSTRYGLAAGVFTQNLATANTLMRALRAGTVWINCFDVFDAAIPFGGYKMSGIGREKGIY 521
Query: 377 SLKNYLQVKAVVTPIKNAAWL 315
SL NYLQVKAVVTP+KN AWL
Sbjct: 522 SLHNYLQVKAVVTPLKNPAWL 542
>sptr|Q8RUR9|Q8RUR9 Mitochondrial aldehyde dehydrogenase RF2B.
Length = 550
Score = 860 bits (2223), Expect = 0.0
Identities = 417/508 (82%), Positives = 464/508 (91%), Gaps = 5/508 (0%)
Frame = -2
Query: 1823 LQRFSTA-----AAVEEPITPSVHVNYTKLLINGNFVDSASGKTFPTLDPRTGEVIAHVA 1659
L RFSTA AA EEPI P+V V +T+LLINGNFVD+ASGKTFPTLDPRTGEVIA VA
Sbjct: 43 LHRFSTAPASAAAAAEEPIQPAVEVKHTQLLINGNFVDAASGKTFPTLDPRTGEVIARVA 102
Query: 1658 EGDAEDINRAVAAARKAFDEGPWPKMTAYERSRILLRFADLIEKHNDELAALETWDNGKP 1479
EGD+EDI+RAVAAAR+AFDEGPWP+MTAY+R R+LLRFADLIE+H +E+AALETWDNGK
Sbjct: 103 EGDSEDIDRAVAAARRAFDEGPWPRMTAYDRCRVLLRFADLIERHAEEVAALETWDNGKT 162
Query: 1478 YEQAAQIEVPMVARLMRYYAGWADKIHGLIVPADGPHHVQILHEPIGVAGQIIPWNFPLL 1299
QAA EVPMVAR +RYYAGWADKIHGL+ PADG HHVQ+LHEP+GVAGQIIPWNFPLL
Sbjct: 163 LAQAAGAEVPMVARCVRYYAGWADKIHGLVAPADGAHHVQVLHEPVGVAGQIIPWNFPLL 222
Query: 1298 MFAWKVGPALACGNTLVLKTAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTAGAALA 1119
MFAWKVGPALACGNT+VLKTAEQTPLSALY++ LLHEAGLPEGV+NVVSGFGPTAGAAL+
Sbjct: 223 MFAWKVGPALACGNTVVLKTAEQTPLSALYVANLLHEAGLPEGVLNVVSGFGPTAGAALS 282
Query: 1118 SHMDVDKIAFTGSTDTGKIILELAAKSNLKTVTLELGGKSPFIIMDDADVDHAVELAHFA 939
SHM VDK+AFTGST TG+I+LELAA+SNLK VTLELGGKSPFI+MDDADVD AVELAH A
Sbjct: 283 SHMGVDKLAFTGSTGTGQIVLELAARSNLKPVTLELGGKSPFIVMDDADVDQAVELAHQA 342
Query: 938 LFFNQGQCCCAGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDEQFNKI 759
+FFNQGQCCCAGSRTFVHERVYDEFVEK+KARALKRVVGDPFR GVEQGPQID EQFNKI
Sbjct: 343 VFFNQGQCCCAGSRTFVHERVYDEFVEKSKARALKRVVGDPFRDGVEQGPQIDGEQFNKI 402
Query: 758 LRYIRYGVDGGATLVTGGDRLGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSILKFKDL 579
LRY++ GVD GATLV GGDR+GD+GFYIQPT+F+D +D MKIA+EEIFGPVQ+ILKF +
Sbjct: 403 LRYVQSGVDSGATLVAGGDRVGDRGFYIQPTVFADAKDEMKIAREEIFGPVQTILKFSGV 462
Query: 578 NEVIKRANASQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGYKMSGI 399
EVI+RANA+ YGLAAGVFT SLD ANTL+RALRAGTVWVNC+DVFDA IPFGGYKMSG+
Sbjct: 463 EEVIRRANATPYGLAAGVFTRSLDAANTLSRALRAGTVWVNCYDVFDATIPFGGYKMSGV 522
Query: 398 GREKGVDSLKNYLQVKAVVTPIKNAAWL 315
GREKG+ +L+NYLQ KAVVTPIKN AWL
Sbjct: 523 GREKGIYALRNYLQTKAVVTPIKNPAWL 550
>sptr|Q8LST5|Q8LST5 Mitochondrial aldehyde dehydrogenase.
Length = 551
Score = 860 bits (2221), Expect = 0.0
Identities = 416/510 (81%), Positives = 463/510 (90%), Gaps = 5/510 (0%)
Frame = -2
Query: 1829 GVLQRFSTA-----AAVEEPITPSVHVNYTKLLINGNFVDSASGKTFPTLDPRTGEVIAH 1665
G L RFSTA A EEPI P+V V +T+LLINGNFVD+ASGKTFPTLDPRTGEVIA
Sbjct: 42 GPLHRFSTAPAAAAATAEEPIQPAVEVKHTQLLINGNFVDAASGKTFPTLDPRTGEVIAR 101
Query: 1664 VAEGDAEDINRAVAAARKAFDEGPWPKMTAYERSRILLRFADLIEKHNDELAALETWDNG 1485
VAEGD+EDI+RAVAAAR+AFDEGPWP+MTAYER R+LLRFADLIE+H +E+AALETWDNG
Sbjct: 102 VAEGDSEDIDRAVAAARRAFDEGPWPRMTAYERCRVLLRFADLIERHAEEIAALETWDNG 161
Query: 1484 KPYEQAAQIEVPMVARLMRYYAGWADKIHGLIVPADGPHHVQILHEPIGVAGQIIPWNFP 1305
K QAA EVPMVAR +RYYAGWADKIHGL+VP DG HHVQ+LHEP+GVAGQIIPWNFP
Sbjct: 162 KTLAQAAGAEVPMVARCIRYYAGWADKIHGLVVPGDGAHHVQVLHEPVGVAGQIIPWNFP 221
Query: 1304 LLMFAWKVGPALACGNTLVLKTAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTAGAA 1125
LLMFAWKVGPALACGNT+VLKTAEQTPLSALY++ LLHEAGLPEGV+NVVSGFGPTAGAA
Sbjct: 222 LLMFAWKVGPALACGNTVVLKTAEQTPLSALYVANLLHEAGLPEGVLNVVSGFGPTAGAA 281
Query: 1124 LASHMDVDKIAFTGSTDTGKIILELAAKSNLKTVTLELGGKSPFIIMDDADVDHAVELAH 945
L SHM VDK+AFTGST TG+I+LELAA+SNLK VTLELGGKSPF++MDDADVD AVELAH
Sbjct: 282 LCSHMGVDKLAFTGSTGTGQIVLELAARSNLKPVTLELGGKSPFVVMDDADVDQAVELAH 341
Query: 944 FALFFNQGQCCCAGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDEQFN 765
A+FFNQGQCCCAGSRTFVHERVYDEFVEK+KARALKRVVGDPFR GVEQGPQID +QFN
Sbjct: 342 QAVFFNQGQCCCAGSRTFVHERVYDEFVEKSKARALKRVVGDPFRNGVEQGPQIDGDQFN 401
Query: 764 KILRYIRYGVDGGATLVTGGDRLGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSILKFK 585
KILRY++ GVD GATLVTGGDR+G +GFYIQPT+F+D +D MKIA+EEIFGPVQ+ILKF
Sbjct: 402 KILRYVQSGVDSGATLVTGGDRVGSRGFYIQPTVFADAKDDMKIAREEIFGPVQTILKFS 461
Query: 584 DLNEVIKRANASQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGYKMS 405
+ EVI+RANA+ YGLAAGVFT SLD ANTL+RALRAGTVWVNC+DVFDA IPFGGYKMS
Sbjct: 462 GMEEVIRRANATHYGLAAGVFTRSLDAANTLSRALRAGTVWVNCYDVFDATIPFGGYKMS 521
Query: 404 GIGREKGVDSLKNYLQVKAVVTPIKNAAWL 315
G+GREKGV +L+NYLQ KAVVTPIK+ AWL
Sbjct: 522 GVGREKGVYALRNYLQTKAVVTPIKDPAWL 551
>sptr|Q9LRI6|Q9LRI6 Mitochondrial aldehyde dehydrogenase ALDH2a.
Length = 553
Score = 859 bits (2219), Expect = 0.0
Identities = 419/514 (81%), Positives = 459/514 (89%), Gaps = 7/514 (1%)
Frame = -2
Query: 1835 LPGVLQRFS-------TAAAVEEPITPSVHVNYTKLLINGNFVDSASGKTFPTLDPRTGE 1677
LPG L RFS TAAA EEPI P V V YTKLLINGNFVD+ASGKTF T+DPRTG+
Sbjct: 40 LPGSLHRFSAAPAAAATAAATEEPIQPPVDVKYTKLLINGNFVDAASGKTFATVDPRTGD 99
Query: 1676 VIAHVAEGDAEDINRAVAAARKAFDEGPWPKMTAYERSRILLRFADLIEKHNDELAALET 1497
VIA VAEGDAED+NRAVAAAR+AFDEGPWP+MTAYER R+LLRFADLIE+H DE+AALET
Sbjct: 100 VIARVAEGDAEDVNRAVAAARRAFDEGPWPRMTAYERCRVLLRFADLIEQHADEIAALET 159
Query: 1496 WDNGKPYEQAAQIEVPMVARLMRYYAGWADKIHGLIVPADGPHHVQILHEPIGVAGQIIP 1317
WD GK EQ EVPMVAR MRYY GWADKIHGL+VPADGPHHVQ+LHEPIGVAGQIIP
Sbjct: 160 WDGGKTLEQTTGTEVPMVARYMRYYGGWADKIHGLVVPADGPHHVQVLHEPIGVAGQIIP 219
Query: 1316 WNFPLLMFAWKVGPALACGNTLVLKTAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPT 1137
WNFPLLMFAWKVGPALACGN +VLKTAEQTPLSAL+++ LLHEAGLP+GV+NVVSGFGPT
Sbjct: 220 WNFPLLMFAWKVGPALACGNAVVLKTAEQTPLSALFVASLLHEAGLPDGVLNVVSGFGPT 279
Query: 1136 AGAALASHMDVDKIAFTGSTDTGKIILELAAKSNLKTVTLELGGKSPFIIMDDADVDHAV 957
AGAAL+SHM VDK+AFTGST TGKI+LELAA+SNLK VTLELGGKSPFI+MDDADVD AV
Sbjct: 280 AGAALSSHMGVDKLAFTGSTGTGKIVLELAARSNLKPVTLELGGKSPFIVMDDADVDQAV 339
Query: 956 ELAHFALFFNQGQCCCAGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDD 777
ELAH ALFFNQGQCCCAGSRTFVHERVYDEFVEKA+ARAL+RVVGDPFR GVEQGPQID
Sbjct: 340 ELAHRALFFNQGQCCCAGSRTFVHERVYDEFVEKARARALQRVVGDPFRTGVEQGPQIDG 399
Query: 776 EQFNKILRYIRYGVDGGATLVTGGDRLGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSI 597
EQF KIL+Y++ GVD GATLV GGDR G +GFYIQPT+F+DV+D MKIAQEEIFGPVQSI
Sbjct: 400 EQFKKILQYVKSGVDSGATLVAGGDRAGSRGFYIQPTVFADVEDEMKIAQEEIFGPVQSI 459
Query: 596 LKFKDLNEVIKRANASQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGG 417
LKF + EV++RANA+ YGLAAGVFT LD ANTL RALR GTVWVN +DVFDAA+PFGG
Sbjct: 460 LKFSTVEEVVRRANATPYGLAAGVFTQRLDAANTLARALRVGTVWVNTYDVFDAAVPFGG 519
Query: 416 YKMSGIGREKGVDSLKNYLQVKAVVTPIKNAAWL 315
YKMSG+GREKGV SL+NYLQ KAVVTPIK+AAWL
Sbjct: 520 YKMSGVGREKGVYSLRNYLQTKAVVTPIKDAAWL 553
>sptr|P93344|P93344 Aldehyde dehydrogenase (NAD+) (EC 1.2.1.3).
Length = 542
Score = 857 bits (2215), Expect = 0.0
Identities = 407/501 (81%), Positives = 459/501 (91%)
Frame = -2
Query: 1817 RFSTAAAVEEPITPSVHVNYTKLLINGNFVDSASGKTFPTLDPRTGEVIAHVAEGDAEDI 1638
++STAAA+EEPI P+V+V +TKL ING FVD+ASGKTFPTLDPRTGEVIAHVAEGDAEDI
Sbjct: 42 QYSTAAAIEEPIKPAVNVEHTKLFINGQFVDAASGKTFPTLDPRTGEVIAHVAEGDAEDI 101
Query: 1637 NRAVAAARKAFDEGPWPKMTAYERSRILLRFADLIEKHNDELAALETWDNGKPYEQAAQI 1458
NRAVAAARKAFDEGPWPKM AYERS+I +R ADLIEKHND++A LETWD GKPYEQAA+I
Sbjct: 102 NRAVAAARKAFDEGPWPKMNAYERSKIFVRLADLIEKHNDQIATLETWDTGKPYEQAAKI 161
Query: 1457 EVPMVARLMRYYAGWADKIHGLIVPADGPHHVQILHEPIGVAGQIIPWNFPLLMFAWKVG 1278
EVPMV RL+RYYAGWADKIHG+ +PADGP+HVQ LHEPIGVAGQIIPWNFPLLMF+WK+G
Sbjct: 162 EVPMVVRLLRYYAGWADKIHGMTIPADGPYHVQTLHEPIGVAGQIIPWNFPLLMFSWKIG 221
Query: 1277 PALACGNTLVLKTAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTAGAALASHMDVDK 1098
PALACGNT+VLKTAEQTPLSA Y++ LL EAGLPEGV+N++SGFGPTAGA L SHMDVDK
Sbjct: 222 PALACGNTVVLKTAEQTPLSAFYVAHLLQEAGLPEGVLNIISGFGPTAGAPLCSHMDVDK 281
Query: 1097 IAFTGSTDTGKIILELAAKSNLKTVTLELGGKSPFIIMDDADVDHAVELAHFALFFNQGQ 918
+AFTGSTDTGK IL LAAKSNLK VTLELGGKSPFI+ +DAD+D AVE AHFALFFNQGQ
Sbjct: 282 LAFTGSTDTGKAILSLAAKSNLKPVTLELGGKSPFIVCEDADIDTAVEQAHFALFFNQGQ 341
Query: 917 CCCAGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDEQFNKILRYIRYG 738
CCCAGSRTFVHE+VYDEF+EKAKARALKR VGDPF+ G EQGPQID +QF+KI+ YIR G
Sbjct: 342 CCCAGSRTFVHEKVYDEFLEKAKARALKRTVGDPFKSGTEQGPQIDSKQFDKIMNYIRSG 401
Query: 737 VDGGATLVTGGDRLGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSILKFKDLNEVIKRA 558
+D GATL TGG+RLG++G+YI+PT+FS+V+D M IAQ+EIFGPVQSILKFKD+++VI+RA
Sbjct: 402 IDSGATLETGGERLGERGYYIKPTVFSNVKDDMLIAQDEIFGPVQSILKFKDVDDVIRRA 461
Query: 557 NASQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGYKMSGIGREKGVD 378
N S+YGLAAGVFT ++DTANTLTRALR GTVWVNCFD FDA IPFGGYKMSG GREKG
Sbjct: 462 NNSRYGLAAGVFTQNIDTANTLTRALRVGTVWVNCFDTFDATIPFGGYKMSGHGREKGEY 521
Query: 377 SLKNYLQVKAVVTPIKNAAWL 315
SLKNYLQVKAVVTP+KN AWL
Sbjct: 522 SLKNYLQVKAVVTPLKNPAWL 542
>sptr|Q94C67|Q94C67 Putative aldehyde dehydrogenase NAD+.
Length = 534
Score = 855 bits (2209), Expect = 0.0
Identities = 413/503 (82%), Positives = 457/503 (90%), Gaps = 1/503 (0%)
Frame = -2
Query: 1820 QRFST-AAAVEEPITPSVHVNYTKLLINGNFVDSASGKTFPTLDPRTGEVIAHVAEGDAE 1644
QR+S AAAVE ITP V V +T+LLI G FVD+ SGKTFPTLDPR GEVIA V+EGDAE
Sbjct: 32 QRYSNLAAAVENTITPPVKVEHTQLLIGGRFVDAVSGKTFPTLDPRNGEVIAQVSEGDAE 91
Query: 1643 DINRAVAAARKAFDEGPWPKMTAYERSRILLRFADLIEKHNDELAALETWDNGKPYEQAA 1464
D+NRAVAAARKAFDEGPWPKMTAYERS+IL RFADLIEKHNDE+AALETWDNGKPYEQ+A
Sbjct: 92 DVNRAVAAARKAFDEGPWPKMTAYERSKILFRFADLIEKHNDEIAALETWDNGKPYEQSA 151
Query: 1463 QIEVPMVARLMRYYAGWADKIHGLIVPADGPHHVQILHEPIGVAGQIIPWNFPLLMFAWK 1284
QIEVPM+AR+ RYYAGWADKIHG+ +P DGPHHVQ LHEPIGVAGQIIPWNFPLLM +WK
Sbjct: 152 QIEVPMLARVFRYYAGWADKIHGMTMPGDGPHHVQTLHEPIGVAGQIIPWNFPLLMLSWK 211
Query: 1283 VGPALACGNTLVLKTAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTAGAALASHMDV 1104
+GPALACGNT+VLKTAEQTPLSAL + KLLHEAGLP+GVVN+VSGFG TAGAA+ASHMDV
Sbjct: 212 LGPALACGNTVVLKTAEQTPLSALLVGKLLHEAGLPDGVVNIVSGFGATAGAAIASHMDV 271
Query: 1103 DKIAFTGSTDTGKIILELAAKSNLKTVTLELGGKSPFIIMDDADVDHAVELAHFALFFNQ 924
DK+AFTGSTD GKIILELA+KSNLK VTLELGGKSPFI+ +DADVD AVELAHFALFFNQ
Sbjct: 272 DKVAFTGSTDVGKIILELASKSNLKAVTLELGGKSPFIVCEDADVDQAVELAHFALFFNQ 331
Query: 923 GQCCCAGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDEQFNKILRYIR 744
GQCCCAGSRTFVHERVYDEFVEKAKARALKR VGDPF+ G+EQGPQ+D EQFNKIL+YI+
Sbjct: 332 GQCCCAGSRTFVHERVYDEFVEKAKARALKRNVGDPFKSGIEQGPQVDSEQFNKILKYIK 391
Query: 743 YGVDGGATLVTGGDRLGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSILKFKDLNEVIK 564
+GV+ GATL GGDRLG KG+YIQPT+FSDV+D M IA +EIFGPVQ+ILKFKDL+EVI
Sbjct: 392 HGVEAGATLQAGGDRLGSKGYYIQPTVFSDVKDDMLIATDEIFGPVQTILKFKDLDEVIA 451
Query: 563 RANASQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGYKMSGIGREKG 384
RAN S+YGLAAGVFT +LDTA+ L RALR GTVW+NCFDV DA+IPFGGYKMSGIGREKG
Sbjct: 452 RANNSRYGLAAGVFTQNLDTAHRLMRALRVGTVWINCFDVLDASIPFGGYKMSGIGREKG 511
Query: 383 VDSLKNYLQVKAVVTPIKNAAWL 315
+ SL NYLQVKAVVT +KN AWL
Sbjct: 512 IYSLNNYLQVKAVVTSLKNPAWL 534
>sptr|Q9ZUB6|Q9ZUB6 F5O8.35 protein.
Length = 519
Score = 847 bits (2187), Expect = 0.0
Identities = 413/514 (80%), Positives = 457/514 (88%), Gaps = 12/514 (2%)
Frame = -2
Query: 1820 QRFST-AAAVEEPITPSVHVNYTKLLINGNFVDSASGKTFPTLDPRTGEVIAHVAEGDAE 1644
QR+S AAAVE ITP V V +T+LLI G FVD+ SGKTFPTLDPR GEVIA V+EGDAE
Sbjct: 6 QRYSNLAAAVENTITPPVKVEHTQLLIGGRFVDAVSGKTFPTLDPRNGEVIAQVSEGDAE 65
Query: 1643 DINRAVAAARKAFDEGPWPKMTAY-----------ERSRILLRFADLIEKHNDELAALET 1497
D+NRAVAAARKAFDEGPWPKMTAY ERS+IL RFADLIEKHNDE+AALET
Sbjct: 66 DVNRAVAAARKAFDEGPWPKMTAYVYNKVVLCLSSERSKILFRFADLIEKHNDEIAALET 125
Query: 1496 WDNGKPYEQAAQIEVPMVARLMRYYAGWADKIHGLIVPADGPHHVQILHEPIGVAGQIIP 1317
WDNGKPYEQ+AQIEVPM+AR+ RYYAGWADKIHG+ +P DGPHHVQ LHEPIGVAGQIIP
Sbjct: 126 WDNGKPYEQSAQIEVPMLARVFRYYAGWADKIHGMTMPGDGPHHVQTLHEPIGVAGQIIP 185
Query: 1316 WNFPLLMFAWKVGPALACGNTLVLKTAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPT 1137
WNFPLLM +WK+GPALACGNT+VLKTAEQTPLSAL + KLLHEAGLP+GVVN+VSGFG T
Sbjct: 186 WNFPLLMLSWKLGPALACGNTVVLKTAEQTPLSALLVGKLLHEAGLPDGVVNIVSGFGAT 245
Query: 1136 AGAALASHMDVDKIAFTGSTDTGKIILELAAKSNLKTVTLELGGKSPFIIMDDADVDHAV 957
AGAA+ASHMDVDK+AFTGSTD GKIILELA+KSNLK VTLELGGKSPFI+ +DADVD AV
Sbjct: 246 AGAAIASHMDVDKVAFTGSTDVGKIILELASKSNLKAVTLELGGKSPFIVCEDADVDQAV 305
Query: 956 ELAHFALFFNQGQCCCAGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDD 777
ELAHFALFFNQGQCCCAGSRTFVHERVYDEFVEKAKARALKR VGDPF+ G+EQGPQ+D
Sbjct: 306 ELAHFALFFNQGQCCCAGSRTFVHERVYDEFVEKAKARALKRNVGDPFKSGIEQGPQVDS 365
Query: 776 EQFNKILRYIRYGVDGGATLVTGGDRLGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSI 597
EQFNKIL+YI++GV+ GATL GGDRLG KG+YIQPT+FSDV+D M IA +EIFGPVQ+I
Sbjct: 366 EQFNKILKYIKHGVEAGATLQAGGDRLGSKGYYIQPTVFSDVKDDMLIATDEIFGPVQTI 425
Query: 596 LKFKDLNEVIKRANASQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGG 417
LKFKDL+EVI RAN S+YGLAAGVFT +LDTA+ L RALR GTVW+NCFDV DA+IPFGG
Sbjct: 426 LKFKDLDEVIARANNSRYGLAAGVFTQNLDTAHRLMRALRVGTVWINCFDVLDASIPFGG 485
Query: 416 YKMSGIGREKGVDSLKNYLQVKAVVTPIKNAAWL 315
YKMSGIGREKG+ SL NYLQVKAVVT +KN AWL
Sbjct: 486 YKMSGIGREKGIYSLNNYLQVKAVVTSLKNPAWL 519
>sptr|Q8S528|Q8S528 Mitochondrial aldehyde dehydrogenase.
Length = 534
Score = 837 bits (2162), Expect = 0.0
Identities = 406/503 (80%), Positives = 450/503 (89%), Gaps = 1/503 (0%)
Frame = -2
Query: 1820 QRFST-AAAVEEPITPSVHVNYTKLLINGNFVDSASGKTFPTLDPRTGEVIAHVAEGDAE 1644
QR+S AAAVE ITP V V +T+LLI G FVD+ SGKTFPTLDPR GEVIA V+EGDAE
Sbjct: 32 QRYSNLAAAVENTITPPVKVEHTQLLIGGRFVDAVSGKTFPTLDPRNGEVIAQVSEGDAE 91
Query: 1643 DINRAVAAARKAFDEGPWPKMTAYERSRILLRFADLIEKHNDELAALETWDNGKPYEQAA 1464
D+NRAVAAARKAFDEGPWPKMTAYERS+IL RFADLIEKHNDE+AALETWDNGKPYEQ+A
Sbjct: 92 DVNRAVAAARKAFDEGPWPKMTAYERSKILFRFADLIEKHNDEIAALETWDNGKPYEQSA 151
Query: 1463 QIEVPMVARLMRYYAGWADKIHGLIVPADGPHHVQILHEPIGVAGQIIPWNFPLLMFAWK 1284
QIEVPM+AR+ RYYAGWADKIHG+ +P DGPHHVQ LHEPIGVAGQIIPWNFPLLM +WK
Sbjct: 152 QIEVPMLARVFRYYAGWADKIHGMTMPGDGPHHVQTLHEPIGVAGQIIPWNFPLLMLSWK 211
Query: 1283 VGPALACGNTLVLKTAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTAGAALASHMDV 1104
+GPALACGNT+VLKTAEQTPLSAL + KLLHEAGLP+GVVN+VSGFG TAGAA+ASHMDV
Sbjct: 212 LGPALACGNTVVLKTAEQTPLSALLVGKLLHEAGLPDGVVNIVSGFGATAGAAIASHMDV 271
Query: 1103 DKIAFTGSTDTGKIILELAAKSNLKTVTLELGGKSPFIIMDDADVDHAVELAHFALFFNQ 924
DK+AFTGSTD GKIILELA+KSNLK VTLEL + +DADVD AVELAHFALFFNQ
Sbjct: 272 DKVAFTGSTDVGKIILELASKSNLKAVTLELEESHHSFVCEDADVDQAVELAHFALFFNQ 331
Query: 923 GQCCCAGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDEQFNKILRYIR 744
GQCCCAGSRTFVHERVYDEFVEKAKARALKR VGDPF+ G+EQGPQ+D EQFNKIL+YI+
Sbjct: 332 GQCCCAGSRTFVHERVYDEFVEKAKARALKRNVGDPFKSGIEQGPQVDSEQFNKILKYIK 391
Query: 743 YGVDGGATLVTGGDRLGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSILKFKDLNEVIK 564
+GV+ GATL GGDRLG KG+YIQPT+FSDV+D M IA +EIFGPVQ+ILKFKDL+EVI
Sbjct: 392 HGVEAGATLQAGGDRLGSKGYYIQPTVFSDVKDDMLIATDEIFGPVQTILKFKDLDEVIA 451
Query: 563 RANASQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGYKMSGIGREKG 384
RAN S+YGLAAGVFT +LDTA+ L RALR GTVW+NCFDV DA+IPFGGYKMSGIGREKG
Sbjct: 452 RANNSRYGLAAGVFTQNLDTAHRLMRALRVGTVWINCFDVLDASIPFGGYKMSGIGREKG 511
Query: 383 VDSLKNYLQVKAVVTPIKNAAWL 315
+ SL NYLQVKAVVT +KN AWL
Sbjct: 512 IYSLNNYLQVKAVVTSLKNPAWL 534
>sptr|Q9SU63|Q9SU63 Aldehyde dehydrogenase (NAD+)-like protein (EC
1.2.1.3) (Putative aldehyde dehydrogenase (NAD+) protein)
(AT3g48000/T17F15_130) (Aldehyde dehydrogenase ALDH2a)
(NAD+).
Length = 538
Score = 833 bits (2151), Expect = 0.0
Identities = 400/506 (79%), Positives = 456/506 (90%), Gaps = 1/506 (0%)
Frame = -2
Query: 1829 GVLQRFSTA-AAVEEPITPSVHVNYTKLLINGNFVDSASGKTFPTLDPRTGEVIAHVAEG 1653
G+L+RF T+ AA EE I PSV V++T+LLINGNFVDSASGKTFPTLDPRTGEVIAHVAEG
Sbjct: 33 GILRRFGTSSAAAEEIINPSVQVSHTQLLINGNFVDSASGKTFPTLDPRTGEVIAHVAEG 92
Query: 1652 DAEDINRAVAAARKAFDEGPWPKMTAYERSRILLRFADLIEKHNDELAALETWDNGKPYE 1473
DAEDINRAV AAR AFDEGPWPKM+AYERSR+LLRFADL+EKH++ELA+LETWDNGKPY+
Sbjct: 93 DAEDINRAVKAARTAFDEGPWPKMSAYERSRVLLRFADLVEKHSEELASLETWDNGKPYQ 152
Query: 1472 QAAQIEVPMVARLMRYYAGWADKIHGLIVPADGPHHVQILHEPIGVAGQIIPWNFPLLMF 1293
Q+ E+PM ARL RYYAGWADKIHGL +PADG + V LHEPIGVAGQIIPWNFPLLMF
Sbjct: 153 QSLTAEIPMFARLFRYYAGWADKIHGLTIPADGNYQVHTLHEPIGVAGQIIPWNFPLLMF 212
Query: 1292 AWKVGPALACGNTLVLKTAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTAGAALASH 1113
AWKVGPALACGNT+VLKTAEQTPL+A Y KL EAGLP GV+N+VSGFG TAGAALASH
Sbjct: 213 AWKVGPALACGNTIVLKTAEQTPLTAFYAGKLFLEAGLPPGVLNIVSGFGATAGAALASH 272
Query: 1112 MDVDKIAFTGSTDTGKIILELAAKSNLKTVTLELGGKSPFIIMDDADVDHAVELAHFALF 933
MDVDK+AFTGSTDTGK+IL LAA SNLK VTLELGGKSPFI+ +DAD+D AVELAHFALF
Sbjct: 273 MDVDKLAFTGSTDTGKVILGLAANSNLKPVTLELGGKSPFIVFEDADIDKAVELAHFALF 332
Query: 932 FNQGQCCCAGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDEQFNKILR 753
FNQGQCCCAGSRTFVHE+VYDEFVEK+KARALKRVVGDPFRKG+EQGPQID +QF K+++
Sbjct: 333 FNQGQCCCAGSRTFVHEKVYDEFVEKSKARALKRVVGDPFRKGIEQGPQIDLKQFEKVMK 392
Query: 752 YIRYGVDGGATLVTGGDRLGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSILKFKDLNE 573
YI+ G++ ATL GGD++GDKG++IQPT+FS+V+D M IAQ+EIFGPVQSILKF D++E
Sbjct: 393 YIKSGIESNATLECGGDQIGDKGYFIQPTVFSNVKDDMLIAQDEIFGPVQSILKFSDVDE 452
Query: 572 VIKRANASQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGYKMSGIGR 393
VIKRAN ++YGLAAGVFT +LDTAN ++RAL+AGTVWVNCFDVFDAAIPFGGYKMSG GR
Sbjct: 453 VIKRANETKYGLAAGVFTKNLDTANRVSRALKAGTVWVNCFDVFDAAIPFGGYKMSGNGR 512
Query: 392 EKGVDSLKNYLQVKAVVTPIKNAAWL 315
EKG+ SL NYLQ+KAVVT + AW+
Sbjct: 513 EKGIYSLNNYLQIKAVVTALNKPAWI 538
>sptr|Q8GU27|Q8GU27 Aldehyde dehydrogenase.
Length = 523
Score = 612 bits (1577), Expect = e-173
Identities = 304/507 (59%), Positives = 367/507 (72%), Gaps = 7/507 (1%)
Frame = -2
Query: 1817 RFSTAAAVEEPITP--SVHVNYT-----KLLINGNFVDSASGKTFPTLDPRTGEVIAHVA 1659
RF+ +A+ P+ S + + KL IN +VD+ SGKT DPR GE I VA
Sbjct: 17 RFARSASTSAPVVDIASETIQFARNLPNKLFINNKWVDAKSGKTMGVEDPRNGEEILRVA 76
Query: 1658 EGDAEDINRAVAAARKAFDEGPWPKMTAYERSRILLRFADLIEKHNDELAALETWDNGKP 1479
EGD+ D++ AV AARKAFDEG WP+MT ER R++ + AD +E + + A LET DNGKP
Sbjct: 77 EGDSADVDLAVKAARKAFDEGAWPRMTGKERGRLIYKLADALEANAEAFARLETLDNGKP 136
Query: 1478 YEQAAQIEVPMVARLMRYYAGWADKIHGLIVPADGPHHVQILHEPIGVAGQIIPWNFPLL 1299
+ +VP+ RYYAGWADKIHG +P DGPH LHEP+GV G IIPWNFPLL
Sbjct: 137 LNISRIADVPLSVDHFRYYAGWADKIHGKTIPVDGPHMAYTLHEPLGVVGAIIPWNFPLL 196
Query: 1298 MFAWKVGPALACGNTLVLKTAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTAGAALA 1119
M AWK+GPALA GNT+VLK AEQTPLSAL I+KL E G PEGV+NVV+GFGPT G LA
Sbjct: 197 MAAWKLGPALAAGNTIVLKPAEQTPLSALLIAKLAAEVGFPEGVINVVTGFGPTVGGPLA 256
Query: 1118 SHMDVDKIAFTGSTDTGKIILELAAKSNLKTVTLELGGKSPFIIMDDADVDHAVELAHFA 939
SH VDK AFTGST+ GKI+ LAA+ +K TLELGGKSP I+ D DVD AV AH A
Sbjct: 257 SHKLVDKTAFTGSTEVGKIVARLAAEQ-IKPCTLELGGKSPIIVCPDVDVDRAVADAHMA 315
Query: 938 LFFNQGQCCCAGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDEQFNKI 759
LFFN GQCC AGSR +VHE++YDEFVEKA A VGDPF +QGPQ+D+ QF+KI
Sbjct: 316 LFFNHGQCCAAGSRVYVHEKIYDEFVEKATKAAANHSVGDPFSGQYDQGPQVDNAQFSKI 375
Query: 758 LRYIRYGVDGGATLVTGGDRLGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSILKFKDL 579
L Y+ +G GA L GG R+G+KG+Y+ PT+FS+V D MKIA+EEIFGPVQSILK+
Sbjct: 376 LSYVEHGKAEGAKLNVGGCRVGNKGYYVAPTVFSNVTDNMKIAREEIFGPVQSILKYSTF 435
Query: 578 NEVIKRANASQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGYKMSGI 399
+EVI+RANAS YGLA+GVF+ LDT NTL R L AGTVWVNC+++FD+A+PFGG+K SGI
Sbjct: 436 DEVIRRANASDYGLASGVFSKDLDTVNTLVRGLHAGTVWVNCYNLFDSAVPFGGFKTSGI 495
Query: 398 GREKGVDSLKNYLQVKAVVTPIKNAAW 318
GREKG +L NY +VKAV P+ N AW
Sbjct: 496 GREKGEYALSNYTKVKAVYMPLVNPAW 522
>sw|P20000|DHAM_BOVIN Aldehyde dehydrogenase, mitochondrial precursor
(EC 1.2.1.3) (ALDH class 2) (ALDHI) (ALDH-E2).
Length = 520
Score = 611 bits (1576), Expect = e-173
Identities = 312/511 (61%), Positives = 377/511 (73%), Gaps = 5/511 (0%)
Frame = -2
Query: 1841 RLLPGVLQRFSTAAAVEEPITPSVH--VNYTKLLINGNFVDSASGKTFPTLDPRTGEVIA 1668
RL P +R +AA P TP+ V Y ++ IN + D+ S KTFPT++P TG+VI
Sbjct: 11 RLGPRQGRRLLSAATQAVP-TPNQQPEVLYNQIFINNEWHDAVSKKTFPTVNPSTGDVIC 69
Query: 1667 HVAEGDAEDINRAVAAARKAFDEG-PWPKMTAYERSRILLRFADLIEKHNDELAALETWD 1491
HVAEGD D++RAV AAR AF G PW +M A ER R+L R ADLIE+ LAALET D
Sbjct: 70 HVAEGDKADVDRAVKAARAAFQLGSPWRRMDASERGRLLNRLADLIERDRTYLAALETLD 129
Query: 1490 NGKPYEQAAQIEVPMVARLMRYYAGWADKIHGLIVPADGPHHVQILHEPIGVAGQIIPWN 1311
NGKPY + +++ MV + +RYYAGWADK HG +P DG + HEP+GV GQIIPWN
Sbjct: 130 NGKPYIISYLVDLDMVLKCLRYYAGWADKYHGKTIPIDGDYFSYTRHEPVGVCGQIIPWN 189
Query: 1310 FPLLMFAWKVGPALACGNTLVLKTAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTAG 1131
FPLLM AWK+GPALA GN +V+K AEQTPL+ALY++ L+ EAG P GVVNV+ GFGPTAG
Sbjct: 190 FPLLMQAWKLGPALATGNVVVMKVAEQTPLTALYVANLIKEAGFPPGVVNVIPGFGPTAG 249
Query: 1130 AALASHMDVDKIAFTGSTDTGKIILELAAKSNLKTVTLELGGKSPFIIMDDADVDHAVEL 951
AA+ASH DVDK+AFTGST+ G +I A KSNLK VTLE+GGKSP IIM DAD+D AVE
Sbjct: 250 AAIASHEDVDKVAFTGSTEVGHLIQVAAGKSNLKRVTLEIGGKSPNIIMSDADMDWAVEQ 309
Query: 950 AHFALFFNQGQCCCAGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDEQ 771
AHFALFFNQGQCCCAGSRTFV E +Y EFVE++ ARA RVVG+PF EQGPQ+D+ Q
Sbjct: 310 AHFALFFNQGQCCCAGSRTFVQEDIYAEFVERSVARAKSRVVGNPFDSRTEQGPQVDETQ 369
Query: 770 FNKILRYIRYGVDGGATLVTGGDRLGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSILK 591
F K+L YI+ G + G L+ GG D+G++IQPT+F D+QDGM IA+EEIFGPV ILK
Sbjct: 370 FKKVLGYIKSGKEEGLKLLCGGGAAADRGYFIQPTVFGDLQDGMTIAKEEIFGPVMQILK 429
Query: 590 FKDLNEVIKRANASQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGYK 411
FK + EV+ RAN S+YGLAA VFT LD AN L++AL+AGTVWVNC+DVF A PFGGYK
Sbjct: 430 FKSMEEVVGRANNSKYGLAAAVFTKDLDKANYLSQALQAGTVWVNCYDVFGAQSPFGGYK 489
Query: 410 MSGIGREKGVDSLKNYLQVKAVV--TPIKNA 324
+SG GRE G L+ Y +VK V P KN+
Sbjct: 490 LSGSGRELGEYGLQAYTEVKTVTVRVPQKNS 520
>sw|P05091|DHAM_HUMAN Aldehyde dehydrogenase, mitochondrial precursor
(EC 1.2.1.3) (ALDH class 2) (ALDHI) (ALDH-E2).
Length = 517
Score = 610 bits (1573), Expect = e-173
Identities = 313/511 (61%), Positives = 373/511 (72%), Gaps = 5/511 (0%)
Frame = -2
Query: 1841 RLLPGVLQRFSTAAAVEEPITPSVH--VNYTKLLINGNFVDSASGKTFPTLDPRTGEVIA 1668
R P + +R +AAA + P+ V ++ IN + D+ S KTFPT++P TGEVI
Sbjct: 7 RFGPRLGRRLLSAAATQAVPAPNQQPEVFCNQIFINNEWHDAVSRKTFPTVNPSTGEVIC 66
Query: 1667 HVAEGDAEDINRAVAAARKAFDEG-PWPKMTAYERSRILLRFADLIEKHNDELAALETWD 1491
VAEGD ED+++AV AAR AF G PW +M A R R+L R ADLIE+ LAALET D
Sbjct: 67 QVAEGDKEDVDKAVKAARAAFQLGSPWRRMDASHRGRLLNRLADLIERDRTYLAALETLD 126
Query: 1490 NGKPYEQAAQIEVPMVARLMRYYAGWADKIHGLIVPADGPHHVQILHEPIGVAGQIIPWN 1311
NGKPY + +++ MV + +RYYAGWADK HG +P DG HEP+GV GQIIPWN
Sbjct: 127 NGKPYVISYLVDLDMVLKCLRYYAGWADKYHGKTIPIDGDFFSYTRHEPVGVCGQIIPWN 186
Query: 1310 FPLLMFAWKVGPALACGNTLVLKTAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTAG 1131
FPLLM AWK+GPALA GN +V+K AEQTPL+ALY++ L+ EAG P GVVN+V GFGPTAG
Sbjct: 187 FPLLMQAWKLGPALATGNVVVMKVAEQTPLTALYVANLIKEAGFPPGVVNIVPGFGPTAG 246
Query: 1130 AALASHMDVDKIAFTGSTDTGKIILELAAKSNLKTVTLELGGKSPFIIMDDADVDHAVEL 951
AA+ASH DVDK+AFTGST+ G++I A SNLK VTLELGGKSP IIM DAD+D AVE
Sbjct: 247 AAIASHEDVDKVAFTGSTEIGRVIQVAAGSSNLKRVTLELGGKSPNIIMSDADMDWAVEQ 306
Query: 950 AHFALFFNQGQCCCAGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDEQ 771
AHFALFFNQGQCCCAGSRTFV E +YDEFVE++ ARA RVVG+PF EQGPQ+D+ Q
Sbjct: 307 AHFALFFNQGQCCCAGSRTFVQEDIYDEFVERSVARAKSRVVGNPFDSKTEQGPQVDETQ 366
Query: 770 FNKILRYIRYGVDGGATLVTGGDRLGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSILK 591
F KIL YI G GA L+ GG D+G++IQPT+F DVQDGM IA+EEIFGPV ILK
Sbjct: 367 FKKILGYINTGKQEGAKLLCGGGIAADRGYFIQPTVFGDVQDGMTIAKEEIFGPVMQILK 426
Query: 590 FKDLNEVIKRANASQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGYK 411
FK + EV+ RAN S YGLAA VFT LD AN L++AL+AGTVWVNC+DVF A PFGGYK
Sbjct: 427 FKTIEEVVGRANNSTYGLAAAVFTKDLDKANYLSQALQAGTVWVNCYDVFGAQSPFGGYK 486
Query: 410 MSGIGREKGVDSLKNYLQVKAVV--TPIKNA 324
MSG GRE G L+ Y +VK V P KN+
Sbjct: 487 MSGSGRELGEYGLQAYTEVKTVTVKVPQKNS 517
>sptrnew|AAH62081|AAH62081 Aldehyde dehydrogenase 2.
Length = 519
Score = 607 bits (1565), Expect = e-172
Identities = 309/505 (61%), Positives = 371/505 (73%), Gaps = 5/505 (0%)
Frame = -2
Query: 1823 LQRFSTAAAVEEPITPSVH--VNYTKLLINGNFVDSASGKTFPTLDPRTGEVIAHVAEGD 1650
L R +AAA P+ V ++ IN + D+ S KTFPT++P TGEVI VAEG+
Sbjct: 15 LSRLLSAAATSAVPAPNQQPEVFCNQIFINNEWHDAVSKKTFPTVNPSTGEVICQVAEGN 74
Query: 1649 AEDINRAVAAARKAFDEG-PWPKMTAYERSRILLRFADLIEKHNDELAALETWDNGKPYE 1473
ED+++AV AA+ AF G PW +M A +R R+L R ADLIE+ LAALET DNGKPY
Sbjct: 75 KEDVDKAVKAAQAAFQLGSPWRRMDASDRGRLLYRLADLIERDRTYLAALETLDNGKPYV 134
Query: 1472 QAAQIEVPMVARLMRYYAGWADKIHGLIVPADGPHHVQILHEPIGVAGQIIPWNFPLLMF 1293
+ +++ MV + +RYYAGWADK HG +P DG HEP+GV GQIIPWNFPLLM
Sbjct: 135 ISYLVDLDMVLKCLRYYAGWADKYHGKTIPIDGDFFSYTRHEPVGVCGQIIPWNFPLLMQ 194
Query: 1292 AWKVGPALACGNTLVLKTAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTAGAALASH 1113
AWK+GPALA GN +V+K AEQTPL+ALY++ L+ EAG P GVVN+V GFGPTAGAA+ASH
Sbjct: 195 AWKLGPALATGNVVVMKVAEQTPLTALYVANLIKEAGFPPGVVNIVPGFGPTAGAAIASH 254
Query: 1112 MDVDKIAFTGSTDTGKIILELAAKSNLKTVTLELGGKSPFIIMDDADVDHAVELAHFALF 933
DVDK+AFTGST+ G +I A SNLK VTLELGGKSP IIM DAD+D AVE AHFALF
Sbjct: 255 EDVDKVAFTGSTEVGHLIQVAAGSSNLKRVTLELGGKSPNIIMSDADMDWAVEQAHFALF 314
Query: 932 FNQGQCCCAGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDEQFNKILR 753
FNQGQCCCAGSRTFV E VYDEFVE++ ARA RVVG+PF EQGPQ+D+ QF KIL
Sbjct: 315 FNQGQCCCAGSRTFVQEDVYDEFVERSVARAKSRVVGNPFDSRTEQGPQVDETQFKKILG 374
Query: 752 YIRYGVDGGATLVTGGDRLGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSILKFKDLNE 573
YI+ G GA L+ GG D+G++IQPT+F DV+DGM IA+EEIFGPV ILKFK + E
Sbjct: 375 YIKSGQQEGAKLLCGGGAAADRGYFIQPTVFGDVKDGMTIAKEEIFGPVMQILKFKTIEE 434
Query: 572 VIKRANASQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGYKMSGIGR 393
V+ RAN S+YGLAA VFT LD AN L++AL+AGTVW+NC+DVF A PFGGYKMSG GR
Sbjct: 435 VVGRANNSKYGLAAAVFTKDLDKANYLSQALQAGTVWINCYDVFGAQSPFGGYKMSGSGR 494
Query: 392 EKGVDSLKNYLQVKAVV--TPIKNA 324
E G L+ Y +VK V P KN+
Sbjct: 495 ELGEYGLQAYTEVKTVTVKVPQKNS 519
>sw|P11884|DHAM_RAT Aldehyde dehydrogenase, mitochondrial precursor
(EC 1.2.1.3) (ALDH class 2) (ALDH1) (ALDH-E2).
Length = 519
Score = 607 bits (1565), Expect = e-172
Identities = 309/505 (61%), Positives = 371/505 (73%), Gaps = 5/505 (0%)
Frame = -2
Query: 1823 LQRFSTAAAVEEPITPSVH--VNYTKLLINGNFVDSASGKTFPTLDPRTGEVIAHVAEGD 1650
L R +AAA P+ V ++ IN + D+ S KTFPT++P TGEVI VAEG+
Sbjct: 15 LSRLLSAAATSAVPAPNQQPEVFCNQIFINNEWHDAVSKKTFPTVNPSTGEVICQVAEGN 74
Query: 1649 AEDINRAVAAARKAFDEG-PWPKMTAYERSRILLRFADLIEKHNDELAALETWDNGKPYE 1473
ED+++AV AA+ AF G PW +M A +R R+L R ADLIE+ LAALET DNGKPY
Sbjct: 75 KEDVDKAVKAAQAAFQLGSPWRRMDASDRGRLLYRLADLIERDRTYLAALETLDNGKPYV 134
Query: 1472 QAAQIEVPMVARLMRYYAGWADKIHGLIVPADGPHHVQILHEPIGVAGQIIPWNFPLLMF 1293
+ +++ MV + +RYYAGWADK HG +P DG HEP+GV GQIIPWNFPLLM
Sbjct: 135 ISYLVDLDMVLKCLRYYAGWADKYHGKTIPIDGDFFSYTRHEPVGVCGQIIPWNFPLLMQ 194
Query: 1292 AWKVGPALACGNTLVLKTAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTAGAALASH 1113
AWK+GPALA GN +V+K AEQTPL+ALY++ L+ EAG P GVVN+V GFGPTAGAA+ASH
Sbjct: 195 AWKLGPALATGNVVVMKVAEQTPLTALYVANLIKEAGFPPGVVNIVPGFGPTAGAAIASH 254
Query: 1112 MDVDKIAFTGSTDTGKIILELAAKSNLKTVTLELGGKSPFIIMDDADVDHAVELAHFALF 933
DVDK+AFTGST+ G +I A SNLK VTLELGGKSP IIM DAD+D AVE AHFALF
Sbjct: 255 EDVDKVAFTGSTEVGHLIQVAAGSSNLKRVTLELGGKSPNIIMSDADMDWAVEQAHFALF 314
Query: 932 FNQGQCCCAGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDEQFNKILR 753
FNQGQCCCAGSRTFV E VYDEFVE++ ARA RVVG+PF EQGPQ+D+ QF KIL
Sbjct: 315 FNQGQCCCAGSRTFVQEDVYDEFVERSVARAKSRVVGNPFDSRTEQGPQVDETQFKKILG 374
Query: 752 YIRYGVDGGATLVTGGDRLGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSILKFKDLNE 573
YI+ G GA L+ GG D+G++IQPT+F DV+DGM IA+EEIFGPV ILKFK + E
Sbjct: 375 YIKSGQQEGAKLLCGGGAAADRGYFIQPTVFGDVKDGMTIAKEEIFGPVMQILKFKTIEE 434
Query: 572 VIKRANASQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGYKMSGIGR 393
V+ RAN S+YGLAA VFT LD AN L++AL+AGTVW+NC+DVF A PFGGYKMSG GR
Sbjct: 435 VVGRANNSKYGLAAAVFTKDLDKANYLSQALQAGTVWINCYDVFGAQSPFGGYKMSGSGR 494
Query: 392 EKGVDSLKNYLQVKAVV--TPIKNA 324
E G L+ Y +VK V P KN+
Sbjct: 495 ELGEYGLQAYTEVKTVTVKVPQKNS 519
>sptrnew|BAC28959|BAC28959 12 days embryo embryonic body between
diaphragm region and neck cDNA, RIKEN full-length
enriched library, clone:9430093D07 product:aldehyde
dehydrogenase 2, mitochondrial, full insert sequence.
Length = 519
Score = 607 bits (1564), Expect = e-172
Identities = 309/505 (61%), Positives = 370/505 (73%), Gaps = 5/505 (0%)
Frame = -2
Query: 1823 LQRFSTAAAVEEPITPS--VHVNYTKLLINGNFVDSASGKTFPTLDPRTGEVIAHVAEGD 1650
L R +AAA P+ V ++ IN + D+ S KTFPT++P TGEVI VAEG+
Sbjct: 15 LSRLLSAAATSAVPAPNHQPEVFCNQIFINNEWHDAVSRKTFPTVNPSTGEVICQVAEGN 74
Query: 1649 AEDINRAVAAARKAFDEG-PWPKMTAYERSRILLRFADLIEKHNDELAALETWDNGKPYE 1473
ED+++AV AAR AF G PW +M A +R R+L R ADLIE+ LAALET DNGKPY
Sbjct: 75 KEDVDKAVKAARAAFQLGSPWRRMDASDRGRLLYRLADLIERDRTYLAALETLDNGKPYV 134
Query: 1472 QAAQIEVPMVARLMRYYAGWADKIHGLIVPADGPHHVQILHEPIGVAGQIIPWNFPLLMF 1293
+ +++ MV + +RYYAGWADK HG +P DG HEP+GV GQIIPWNFPLLM
Sbjct: 135 ISYLVDLDMVLKCLRYYAGWADKYHGKTIPIDGDFFSYTRHEPVGVCGQIIPWNFPLLMQ 194
Query: 1292 AWKVGPALACGNTLVLKTAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTAGAALASH 1113
AWK+GPALA GN +V+K AEQTPL+ALY++ L+ EAG P GVVN+V GFGPTAGAA+ASH
Sbjct: 195 AWKLGPALATGNVVVMKVAEQTPLTALYVANLIKEAGFPPGVVNIVPGFGPTAGAAIASH 254
Query: 1112 MDVDKIAFTGSTDTGKIILELAAKSNLKTVTLELGGKSPFIIMDDADVDHAVELAHFALF 933
VDK+AFTGST+ G +I A SNLK VTLELGGKSP IIM DAD+D AVE AHFALF
Sbjct: 255 EGVDKVAFTGSTEVGHLIQVAAGSSNLKRVTLELGGKSPNIIMSDADMDWAVEQAHFALF 314
Query: 932 FNQGQCCCAGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDEQFNKILR 753
FNQGQCCCAGSRTFV E VYDEFVE++ ARA RVVG+PF EQGPQ+D+ QF KIL
Sbjct: 315 FNQGQCCCAGSRTFVQENVYDEFVERSVARAKSRVVGNPFDSRTEQGPQVDETQFKKILG 374
Query: 752 YIRYGVDGGATLVTGGDRLGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSILKFKDLNE 573
YI+ G GA L+ GG D+G++IQPT+F DV+DGM IA+EEIFGPV ILKFK + E
Sbjct: 375 YIKSGQQEGAKLLCGGGAAADRGYFIQPTVFGDVKDGMTIAKEEIFGPVMQILKFKTIEE 434
Query: 572 VIKRANASQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGYKMSGIGR 393
V+ RAN S+YGLAA VFT LD AN L++AL+AGTVW+NC+DVF A PFGGYKMSG GR
Sbjct: 435 VVGRANDSKYGLAAAVFTKDLDKANYLSQALQAGTVWINCYDVFGAQSPFGGYKMSGSGR 494
Query: 392 EKGVDSLKNYLQVKAVV--TPIKNA 324
E G L+ Y +VK V P KN+
Sbjct: 495 ELGEYGLQAYTEVKTVTVKVPQKNS 519
>sptrnew|BAC37697|BAC37697 Adult male spinal cord cDNA, RIKEN
full-length enriched library, clone:A330064L07
product:aldehyde dehydrogenase 2, mitochondrial, full
insert sequence.
Length = 519
Score = 607 bits (1564), Expect = e-172
Identities = 309/505 (61%), Positives = 370/505 (73%), Gaps = 5/505 (0%)
Frame = -2
Query: 1823 LQRFSTAAAVEEPITPS--VHVNYTKLLINGNFVDSASGKTFPTLDPRTGEVIAHVAEGD 1650
L R +AAA P+ V ++ IN + D+ S KTFPT++P TGEVI VAEG+
Sbjct: 15 LSRLLSAAATSAVPAPNHQPEVFCNQIFINNEWHDAVSRKTFPTVNPSTGEVICQVAEGN 74
Query: 1649 AEDINRAVAAARKAFDEG-PWPKMTAYERSRILLRFADLIEKHNDELAALETWDNGKPYE 1473
ED+++AV AAR AF G PW +M A +R R+L R ADLIE+ LAALET DNGKPY
Sbjct: 75 KEDVDKAVKAARAAFQLGSPWRRMDASDRGRLLYRLADLIERDRTYLAALETLDNGKPYV 134
Query: 1472 QAAQIEVPMVARLMRYYAGWADKIHGLIVPADGPHHVQILHEPIGVAGQIIPWNFPLLMF 1293
+ +++ MV + +RYYAGWADK HG +P DG HEP+GV GQIIPWNFPLLM
Sbjct: 135 ISYLVDLDMVLKCLRYYAGWADKYHGKTIPIDGDFFSYTRHEPVGVCGQIIPWNFPLLMQ 194
Query: 1292 AWKVGPALACGNTLVLKTAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTAGAALASH 1113
AWK+GPALA GN +V+K AEQTPL+ALY++ L+ EAG P GVVN+V GFGPTAGAA+ASH
Sbjct: 195 AWKLGPALATGNVVVMKVAEQTPLTALYVANLIKEAGFPPGVVNIVPGFGPTAGAAIASH 254
Query: 1112 MDVDKIAFTGSTDTGKIILELAAKSNLKTVTLELGGKSPFIIMDDADVDHAVELAHFALF 933
VDK+AFTGST+ G +I A SNLK VTLELGGKSP IIM DAD+D AVE AHFALF
Sbjct: 255 EGVDKVAFTGSTEVGHLIQVAAGSSNLKRVTLELGGKSPNIIMSDADMDWAVEQAHFALF 314
Query: 932 FNQGQCCCAGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDEQFNKILR 753
FNQGQCCCAGSRTFV E VYDEFVE++ ARA RVVG+PF EQGPQ+D+ QF KIL
Sbjct: 315 FNQGQCCCAGSRTFVQENVYDEFVERSVARAKSRVVGNPFDSRTEQGPQVDETQFKKILG 374
Query: 752 YIRYGVDGGATLVTGGDRLGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSILKFKDLNE 573
YI+ G GA L+ GG D+G++IQPT+F DV+DGM IA+EEIFGPV ILKFK + E
Sbjct: 375 YIKSGQQEGAKLLCGGGAAADRGYFIQPTVFGDVKDGMTIAKEEIFGPVMQILKFKTIEE 434
Query: 572 VIKRANASQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGYKMSGIGR 393
V+ RAN S+YGLAA VFT LD AN L++AL+AGTVW+NC+DVF A PFGGYKMSG GR
Sbjct: 435 VVGRANDSKYGLAAAVFTKDLDKANYLSQALQAGTVWINCYDVFGAQSPFGGYKMSGSGR 494
Query: 392 EKGVDSLKNYLQVKAVV--TPIKNA 324
E G L+ Y +VK V P KN+
Sbjct: 495 ELGEYGLQAYTEVKTVTVKVPQKNS 519
>sptrnew|BAC31225|BAC31225 3 days neonate thymus cDNA, RIKEN
full-length enriched library, clone:A630082J17
product:aldehyde dehydrogenase 2, mitochondrial, full
insert sequence.
Length = 519
Score = 607 bits (1564), Expect = e-172
Identities = 309/505 (61%), Positives = 370/505 (73%), Gaps = 5/505 (0%)
Frame = -2
Query: 1823 LQRFSTAAAVEEPITPS--VHVNYTKLLINGNFVDSASGKTFPTLDPRTGEVIAHVAEGD 1650
L R +AAA P+ V ++ IN + D+ S KTFPT++P TGEVI VAEG+
Sbjct: 15 LSRLLSAAATSAVPAPNHQPEVFCNQIFINNEWHDAVSRKTFPTVNPSTGEVICQVAEGN 74
Query: 1649 AEDINRAVAAARKAFDEG-PWPKMTAYERSRILLRFADLIEKHNDELAALETWDNGKPYE 1473
ED+++AV AAR AF G PW +M A +R R+L R ADLIE+ LAALET DNGKPY
Sbjct: 75 KEDVDKAVKAARAAFQLGSPWRRMDASDRGRLLYRLADLIERDRTYLAALETLDNGKPYV 134
Query: 1472 QAAQIEVPMVARLMRYYAGWADKIHGLIVPADGPHHVQILHEPIGVAGQIIPWNFPLLMF 1293
+ +++ MV + +RYYAGWADK HG +P DG HEP+GV GQIIPWNFPLLM
Sbjct: 135 ISYLVDLDMVLKCLRYYAGWADKYHGKTIPIDGDFFSYTRHEPVGVCGQIIPWNFPLLMQ 194
Query: 1292 AWKVGPALACGNTLVLKTAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTAGAALASH 1113
AWK+GPALA GN +V+K AEQTPL+ALY++ L+ EAG P GVVN+V GFGPTAGAA+ASH
Sbjct: 195 AWKLGPALATGNVVVMKVAEQTPLTALYVANLIKEAGFPPGVVNIVPGFGPTAGAAIASH 254
Query: 1112 MDVDKIAFTGSTDTGKIILELAAKSNLKTVTLELGGKSPFIIMDDADVDHAVELAHFALF 933
VDK+AFTGST+ G +I A SNLK VTLELGGKSP IIM DAD+D AVE AHFALF
Sbjct: 255 EGVDKVAFTGSTEVGHLIQVAAGSSNLKRVTLELGGKSPNIIMSDADMDWAVEQAHFALF 314
Query: 932 FNQGQCCCAGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDEQFNKILR 753
FNQGQCCCAGSRTFV E VYDEFVE++ ARA RVVG+PF EQGPQ+D+ QF KIL
Sbjct: 315 FNQGQCCCAGSRTFVQENVYDEFVERSVARAKSRVVGNPFDSRTEQGPQVDETQFKKILG 374
Query: 752 YIRYGVDGGATLVTGGDRLGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSILKFKDLNE 573
YI+ G GA L+ GG D+G++IQPT+F DV+DGM IA+EEIFGPV ILKFK + E
Sbjct: 375 YIKSGQQEGAKLLCGGGAAADRGYFIQPTVFGDVKDGMTIAKEEIFGPVMQILKFKTIEE 434
Query: 572 VIKRANASQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGYKMSGIGR 393
V+ RAN S+YGLAA VFT LD AN L++AL+AGTVW+NC+DVF A PFGGYKMSG GR
Sbjct: 435 VVGRANDSKYGLAAAVFTKDLDKANYLSQALQAGTVWINCYDVFGAQSPFGGYKMSGSGR 494
Query: 392 EKGVDSLKNYLQVKAVV--TPIKNA 324
E G L+ Y +VK V P KN+
Sbjct: 495 ELGEYGLQAYTEVKTVTVKVPQKNS 519
>sw|P47738|DHAM_MOUSE Aldehyde dehydrogenase, mitochondrial precursor
(EC 1.2.1.3) (ALDH class 2) (AHD-M1) (ALDHI) (ALDH-E2).
Length = 519
Score = 607 bits (1564), Expect = e-172
Identities = 309/505 (61%), Positives = 370/505 (73%), Gaps = 5/505 (0%)
Frame = -2
Query: 1823 LQRFSTAAAVEEPITPS--VHVNYTKLLINGNFVDSASGKTFPTLDPRTGEVIAHVAEGD 1650
L R +AAA P+ V ++ IN + D+ S KTFPT++P TGEVI VAEG+
Sbjct: 15 LSRLLSAAATSAVPAPNHQPEVFCNQIFINNEWHDAVSRKTFPTVNPSTGEVICQVAEGN 74
Query: 1649 AEDINRAVAAARKAFDEG-PWPKMTAYERSRILLRFADLIEKHNDELAALETWDNGKPYE 1473
ED+++AV AAR AF G PW +M A +R R+L R ADLIE+ LAALET DNGKPY
Sbjct: 75 KEDVDKAVKAARAAFQLGSPWRRMDASDRGRLLYRLADLIERDRTYLAALETLDNGKPYV 134
Query: 1472 QAAQIEVPMVARLMRYYAGWADKIHGLIVPADGPHHVQILHEPIGVAGQIIPWNFPLLMF 1293
+ +++ MV + +RYYAGWADK HG +P DG HEP+GV GQIIPWNFPLLM
Sbjct: 135 ISYLVDLDMVLKCLRYYAGWADKYHGKTIPIDGDFFSYTRHEPVGVCGQIIPWNFPLLMQ 194
Query: 1292 AWKVGPALACGNTLVLKTAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTAGAALASH 1113
AWK+GPALA GN +V+K AEQTPL+ALY++ L+ EAG P GVVN+V GFGPTAGAA+ASH
Sbjct: 195 AWKLGPALATGNVVVMKVAEQTPLTALYVANLIKEAGFPPGVVNIVPGFGPTAGAAIASH 254
Query: 1112 MDVDKIAFTGSTDTGKIILELAAKSNLKTVTLELGGKSPFIIMDDADVDHAVELAHFALF 933
VDK+AFTGST+ G +I A SNLK VTLELGGKSP IIM DAD+D AVE AHFALF
Sbjct: 255 EGVDKVAFTGSTEVGHLIQVAAGSSNLKRVTLELGGKSPNIIMSDADMDWAVEQAHFALF 314
Query: 932 FNQGQCCCAGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDEQFNKILR 753
FNQGQCCCAGSRTFV E VYDEFVE++ ARA RVVG+PF EQGPQ+D+ QF KIL
Sbjct: 315 FNQGQCCCAGSRTFVQENVYDEFVERSVARAKSRVVGNPFDSRTEQGPQVDETQFKKILG 374
Query: 752 YIRYGVDGGATLVTGGDRLGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSILKFKDLNE 573
YI+ G GA L+ GG D+G++IQPT+F DV+DGM IA+EEIFGPV ILKFK + E
Sbjct: 375 YIKSGQQEGAKLLCGGGAAADRGYFIQPTVFGDVKDGMTIAKEEIFGPVMQILKFKTIEE 434
Query: 572 VIKRANASQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGYKMSGIGR 393
V+ RAN S+YGLAA VFT LD AN L++AL+AGTVW+NC+DVF A PFGGYKMSG GR
Sbjct: 435 VVGRANDSKYGLAAAVFTKDLDKANYLSQALQAGTVWINCYDVFGAQSPFGGYKMSGSGR 494
Query: 392 EKGVDSLKNYLQVKAVV--TPIKNA 324
E G L+ Y +VK V P KN+
Sbjct: 495 ELGEYGLQAYTEVKTVTVKVPQKNS 519
>sw|P81178|DHAM_MESAU Aldehyde dehydrogenase, mitochondrial (EC
1.2.1.3) (ALDH class 2) (ALDH1) (ALDH-E2).
Length = 500
Score = 606 bits (1563), Expect = e-172
Identities = 308/499 (61%), Positives = 368/499 (73%), Gaps = 3/499 (0%)
Frame = -2
Query: 1811 STAAAVEEPITPSVHVNYTKLLINGNFVDSASGKTFPTLDPRTGEVIAHVAEGDAEDINR 1632
+T+A P V N ++ IN + D+ S KTFPT++P TGEVI VAEG ED+++
Sbjct: 4 ATSAVPAPNQQPEVFCN--QIFINNEWHDAVSKKTFPTVNPSTGEVICQVAEGSKEDVDK 61
Query: 1631 AVAAARKAFDEG-PWPKMTAYERSRILLRFADLIEKHNDELAALETWDNGKPYEQAAQIE 1455
AV AAR AF G PW +M A +R R+L R ADLIE+ LAALET DNGKPY + ++
Sbjct: 62 AVKAARAAFQLGSPWRRMDASDRGRLLNRLADLIERDRTYLAALETLDNGKPYVISYLVD 121
Query: 1454 VPMVARLMRYYAGWADKIHGLIVPADGPHHVQILHEPIGVAGQIIPWNFPLLMFAWKVGP 1275
+ MV + +RYYAGWADK HG +P DG HEP+GV GQIIPWNFPLLM AWK+GP
Sbjct: 122 LDMVLKCLRYYAGWADKYHGKTIPIDGDFFSYTRHEPVGVCGQIIPWNFPLLMQAWKLGP 181
Query: 1274 ALACGNTLVLKTAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTAGAALASHMDVDKI 1095
ALA GN +V+K AEQTPL+ALY++ L+ EAG P GVVN+V GFGPTAGAA+ASH DVDK+
Sbjct: 182 ALATGNVVVMKVAEQTPLTALYVANLIKEAGFPPGVVNIVPGFGPTAGAAIASHEDVDKV 241
Query: 1094 AFTGSTDTGKIILELAAKSNLKTVTLELGGKSPFIIMDDADVDHAVELAHFALFFNQGQC 915
AFTGST+ G +I A SNLK VTLELGGKSP IIM DAD+D AVE AHFALFFNQGQC
Sbjct: 242 AFTGSTEVGHLIQVAAGSSNLKRVTLELGGKSPNIIMSDADMDWAVEQAHFALFFNQGQC 301
Query: 914 CCAGSRTFVHERVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDEQFNKILRYIRYGV 735
CCAGSRTFV E VYDEFVE++ ARA RVVG+PF EQGPQ+D+ QF KIL YI+ G
Sbjct: 302 CCAGSRTFVQEDVYDEFVERSVARAKSRVVGNPFDSRTEQGPQVDETQFKKILGYIKSGQ 361
Query: 734 DGGATLVTGGDRLGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSILKFKDLNEVIKRAN 555
GA L+ GG D+G++IQPT+F DV+DGM IA+EEIFGPV ILKFK + EV+ RAN
Sbjct: 362 QEGAKLLCGGGAAADRGYFIQPTVFGDVKDGMTIAKEEIFGPVMQILKFKTIEEVVGRAN 421
Query: 554 ASQYGLAAGVFTNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGYKMSGIGREKGVDS 375
S+YGLAA VFT LD AN L++AL+AGTVW+NC+DVF A PFGGYKMSG GRE G
Sbjct: 422 NSKYGLAAAVFTKDLDKANYLSQALQAGTVWINCYDVFGAQSPFGGYKMSGSGRELGEYG 481
Query: 374 LKNYLQVKAVV--TPIKNA 324
L+ Y +VK V P KN+
Sbjct: 482 LQAYTEVKTVTIKVPQKNS 500
>sptr|Q8K3V8|Q8K3V8 Mitochondrial aldehyde dehydrogenase (Fragment).
Length = 488
Score = 605 bits (1561), Expect = e-172
Identities = 305/488 (62%), Positives = 365/488 (74%), Gaps = 3/488 (0%)
Frame = -2
Query: 1778 PSVHVNYTKLLINGNFVDSASGKTFPTLDPRTGEVIAHVAEGDAEDINRAVAAARKAFDE 1599
P V N ++ IN + D+ S KTFPT++P TGEVI VAEG+ ED+++AV AA+ AF
Sbjct: 3 PEVFCN--QIFINNEWHDAVSKKTFPTVNPSTGEVICQVAEGNKEDVDKAVKAAQAAFQL 60
Query: 1598 G-PWPKMTAYERSRILLRFADLIEKHNDELAALETWDNGKPYEQAAQIEVPMVARLMRYY 1422
G PW +M A +R R+L R ADLIE+ LAALET DNGKPY + +++ MV + +RYY
Sbjct: 61 GSPWRRMDASDRGRLLYRLADLIERDRTYLAALETLDNGKPYVISYLVDLDMVLKCLRYY 120
Query: 1421 AGWADKIHGLIVPADGPHHVQILHEPIGVAGQIIPWNFPLLMFAWKVGPALACGNTLVLK 1242
AGWADK HG +P DG HEP+GV GQIIPWNFPLLM AWK+GPALA GN +V+K
Sbjct: 121 AGWADKYHGKTIPIDGDFFSYTRHEPVGVCGQIIPWNFPLLMQAWKLGPALATGNVVVMK 180
Query: 1241 TAEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTAGAALASHMDVDKIAFTGSTDTGKI 1062
AEQTPL+ALY++ L+ EAG P GVVN+V GFGPTAGAA+ASH DVDK+AFTGST+ G +
Sbjct: 181 VAEQTPLTALYVANLIKEAGFPPGVVNIVPGFGPTAGAAIASHEDVDKVAFTGSTEVGHL 240
Query: 1061 ILELAAKSNLKTVTLELGGKSPFIIMDDADVDHAVELAHFALFFNQGQCCCAGSRTFVHE 882
I A SNLK VTLELGGKSP IIM DAD+D AVE AHFALFFNQGQCCCAGSRTFV E
Sbjct: 241 IQVAAGSSNLKRVTLELGGKSPNIIMSDADMDWAVEQAHFALFFNQGQCCCAGSRTFVQE 300
Query: 881 RVYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDEQFNKILRYIRYGVDGGATLVTGGD 702
VYDEFVE++ ARA RVVG+PF EQGPQ+D+ QF KIL YI+ G GA L+ GG
Sbjct: 301 DVYDEFVERSVARAKSRVVGNPFDSRTEQGPQVDETQFKKILGYIKSGQQEGAKLLCGGG 360
Query: 701 RLGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSILKFKDLNEVIKRANASQYGLAAGVF 522
D+G++IQPT+F DV+DGM IA+EEIFGPV ILKFK + EV+ RAN S+YGLAA VF
Sbjct: 361 AAADRGYFIQPTVFGDVKDGMTIAKEEIFGPVMQILKFKTIEEVVGRANNSKYGLAAAVF 420
Query: 521 TNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGYKMSGIGREKGVDSLKNYLQVKAVV 342
T LD AN L++AL+AGTVW+NC+DVF A PFGGYKMSG GRE G L+ Y +VK V
Sbjct: 421 TKDLDKANYLSQALQAGTVWINCYDVFGAQSPFGGYKMSGSGRELGEYGLQAYTEVKTVT 480
Query: 341 --TPIKNA 324
P KN+
Sbjct: 481 VKVPQKNS 488
>sptr|Q8S532|Q8S532 Cytosolic aldehyde dehydrogenase RF2C.
Length = 502
Score = 605 bits (1559), Expect = e-171
Identities = 289/488 (59%), Positives = 370/488 (75%)
Frame = -2
Query: 1778 PSVHVNYTKLLINGNFVDSASGKTFPTLDPRTGEVIAHVAEGDAEDINRAVAAARKAFDE 1599
P V V +TKL I+G FVD+ SGKTF T DPRTGEVIA +AEG D++ AV AAR+AFD
Sbjct: 15 PKVEVRFTKLFIDGKFVDAVSGKTFETRDPRTGEVIASIAEGGKADVDLAVKAAREAFDN 74
Query: 1598 GPWPKMTAYERSRILLRFADLIEKHNDELAALETWDNGKPYEQAAQIEVPMVARLMRYYA 1419
GPWP+MT YER RIL RFADLI++H +ELAAL+T D GK + ++P A L+RYYA
Sbjct: 75 GPWPRMTGYERGRILHRFADLIDEHVEELAALDTVDAGKLFAVGKARDIPGAAHLLRYYA 134
Query: 1418 GWADKIHGLIVPADGPHHVQILHEPIGVAGQIIPWNFPLLMFAWKVGPALACGNTLVLKT 1239
G ADK+HG + H L EP+GV G I+PWN+P MF +KVGPALA G +V+K
Sbjct: 135 GAADKVHGATLKMAQRMHGYTLKEPVGVVGHIVPWNYPTTMFFFKVGPALAAGCAVVVKP 194
Query: 1238 AEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTAGAALASHMDVDKIAFTGSTDTGKII 1059
AEQTPLSAL+ + L EAG+P GV+NVV GFGPTAGAA+A+HMDVDK++FTGST+ G+++
Sbjct: 195 AEQTPLSALFYAHLAREAGVPAGVLNVVPGFGPTAGAAVAAHMDVDKVSFTGSTEVGRLV 254
Query: 1058 LELAAKSNLKTVTLELGGKSPFIIMDDADVDHAVELAHFALFFNQGQCCCAGSRTFVHER 879
+ AA+SNLK V+LELGGKSP I+ DDAD+D AV L +FA + N+G+ C AG+R +V E
Sbjct: 255 MRAAAESNLKPVSLELGGKSPVIVFDDADLDMAVNLVNFATYTNKGEICVAGTRIYVQEG 314
Query: 878 VYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDEQFNKILRYIRYGVDGGATLVTGGDR 699
+YDEFV+KA A K VVGDPF V QGPQ+D +Q+ K+LRYI G GATLVTGG
Sbjct: 315 IYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKREGATLVTGGKP 374
Query: 698 LGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSILKFKDLNEVIKRANASQYGLAAGVFT 519
GDKG+YI+PTIF+DV+D M IAQ+EIFGPV +++KFK + EVI++AN ++YGLAAG+ T
Sbjct: 375 CGDKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKANNTRYGLAAGIVT 434
Query: 518 NSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGYKMSGIGREKGVDSLKNYLQVKAVVT 339
++D ANT++R++RAG +W+NC+ FD PFGGYKMSG G++ G+D+L YLQ K VVT
Sbjct: 435 KNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDALDKYLQTKTVVT 494
Query: 338 PIKNAAWL 315
P+ N WL
Sbjct: 495 PLYNTPWL 502
>sptr|Q9LRE9|Q9LRE9 Cytosolic aldehyde dehydrogenase.
Length = 502
Score = 601 bits (1550), Expect = e-170
Identities = 285/488 (58%), Positives = 371/488 (76%)
Frame = -2
Query: 1778 PSVHVNYTKLLINGNFVDSASGKTFPTLDPRTGEVIAHVAEGDAEDINRAVAAARKAFDE 1599
P + + +TKL ING FVD+ SGKTF T DPRTGEVIA +AEGD DI+ AV AAR+AFD
Sbjct: 15 PKLEIKFTKLFINGRFVDAVSGKTFETRDPRTGEVIAKIAEGDKADIDLAVKAAREAFDH 74
Query: 1598 GPWPKMTAYERSRILLRFADLIEKHNDELAALETWDNGKPYEQAAQIEVPMVARLMRYYA 1419
GPWP+M+ + R RIL +FADL+E+H +ELAAL+T D GK + +++P A L+RYYA
Sbjct: 75 GPWPRMSGFARGRILHKFADLVEQHVEELAALDTVDAGKLFAMGKLVDIPGGANLLRYYA 134
Query: 1418 GWADKIHGLIVPADGPHHVQILHEPIGVAGQIIPWNFPLLMFAWKVGPALACGNTLVLKT 1239
G ADK+HG + P H L EP+GV G I+PWN+P MF +K PALA G T+V+K
Sbjct: 135 GAADKVHGETLKMARPCHGYTLKEPVGVVGHIVPWNYPTTMFFFKASPALAAGCTMVVKP 194
Query: 1238 AEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTAGAALASHMDVDKIAFTGSTDTGKII 1059
AEQTPLSAL+ + L AG+P+GV+NVV GFGPTAGAA++SHMD+DK++FTGST+ G+++
Sbjct: 195 AEQTPLSALFYAHLAKLAGVPDGVLNVVPGFGPTAGAAISSHMDIDKVSFTGSTEVGRLV 254
Query: 1058 LELAAKSNLKTVTLELGGKSPFIIMDDADVDHAVELAHFALFFNQGQCCCAGSRTFVHER 879
+E AAKSNLK V+LELGGKSP I+ DDAD+D AV L H A + N+G+ C AGSR +V E
Sbjct: 255 MEAAAKSNLKPVSLELGGKSPVIVFDDADLDTAVNLVHMASYTNKGEICVAGSRIYVQEG 314
Query: 878 VYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDEQFNKILRYIRYGVDGGATLVTGGDR 699
+YD FV+KA A K VVGDPF V QGPQID EQ+ KIL+YI G GATLVTGG
Sbjct: 315 IYDAFVKKATEMAKKSVVGDPFNPRVHQGPQIDKEQYEKILKYIDIGKREGATLVTGGKP 374
Query: 698 LGDKGFYIQPTIFSDVQDGMKIAQEEIFGPVQSILKFKDLNEVIKRANASQYGLAAGVFT 519
G+ G+YI+PTIF+DV++ M IAQEEIFGPV +++KFK + E I++AN+++YGLAAG+ T
Sbjct: 375 CGENGYYIEPTIFTDVKEEMSIAQEEIFGPVMALMKFKTVEEAIQKANSTRYGLAAGIVT 434
Query: 518 NSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGYKMSGIGREKGVDSLKNYLQVKAVVT 339
++D ANT++R++RAG +W+NC+ FD +PFGGYKMSG G++ G+D+L+ YL KAVVT
Sbjct: 435 KNIDVANTVSRSIRAGAIWINCYLGFDPDVPFGGYKMSGFGKDMGMDALEKYLHTKAVVT 494
Query: 338 PIKNAAWL 315
P+ N WL
Sbjct: 495 PLYNTPWL 502
>sptr|Q8S531|Q8S531 Cytosolic aldehyde dehydrogenase RF2C.
Length = 503
Score = 600 bits (1547), Expect = e-170
Identities = 289/489 (59%), Positives = 370/489 (75%), Gaps = 1/489 (0%)
Frame = -2
Query: 1778 PSVHVNYTKLLINGNFVDSASGKTFPTLDPRTGEVIAHVAEGDAEDINRAVAAARKAFDE 1599
P V V +TKL I+G FVD+ SGKTF T DPRTGEVIA +AEG D++ AV AAR+AFD
Sbjct: 15 PKVEVRFTKLFIDGKFVDAVSGKTFETRDPRTGEVIASIAEGGKADVDLAVKAAREAFDN 74
Query: 1598 GPWPKMTAYERSRILLRFADLIEKHNDELAALETWDNGKPYEQAAQIEVPMVARLMRYYA 1419
GPWP+MT YER RIL RFADLI++H +ELAAL+T D GK + ++P A L+RYYA
Sbjct: 75 GPWPRMTGYERGRILHRFADLIDEHVEELAALDTVDAGKLFAVGKARDIPGAAHLLRYYA 134
Query: 1418 GWADKIHGLIVPADGPHHVQILHEPIGVAGQIIPWNFPLLMFAWKVGPALACGNTLVLKT 1239
G ADK+HG + H L EP+GV G I+PWN+P MF +KVGPALA G +V+K
Sbjct: 135 GAADKVHGATLKMAQRMHGYTLKEPVGVVGHIVPWNYPTTMFFFKVGPALAAGCAVVVKP 194
Query: 1238 AEQTPLSALYISKLLHEAGLPEGVVNVVSGFGPTAGAALASHMDVDKIAFTGSTDTGKII 1059
AEQTPLSAL+ + L EAG+P GV+NVV GFGPTAGAA+A+HMDVDK++FTGST+ G+++
Sbjct: 195 AEQTPLSALFYAHLAREAGVPAGVLNVVPGFGPTAGAAVAAHMDVDKVSFTGSTEVGRLV 254
Query: 1058 LELAAKSNLKTVTLELGGKSPFIIMDDADVDHAVELAHFALFFNQGQCCCAGSRTFVHER 879
+ AA+SNLK V+LELGGKSP I+ DDAD+D AV L +FA + N+G+ C AG+R +V E
Sbjct: 255 MRAAAESNLKPVSLELGGKSPVIVFDDADLDMAVNLVNFATYTNKGEICVAGTRIYVQEG 314
Query: 878 VYDEFVEKAKARALKRVVGDPFRKGVEQGPQIDDEQFNKILRYIRYGVDGGATLVTGGDR 699
+YDEFV+KA A K VVGDPF V QGPQ+D +Q+ K+LRYI G GATLVTGG
Sbjct: 315 IYDEFVKKAAELASKSVVGDPFNPSVSQGPQVDKDQYEKVLRYIDIGKREGATLVTGGKP 374
Query: 698 LGD-KGFYIQPTIFSDVQDGMKIAQEEIFGPVQSILKFKDLNEVIKRANASQYGLAAGVF 522
GD KG+YI+PTIF+DV+D M IAQ+EIFGPV +++KFK + EVI++AN ++YGLAAG+
Sbjct: 375 CGDNKGYYIEPTIFTDVKDDMTIAQDEIFGPVMALMKFKTVEEVIQKANNTRYGLAAGIV 434
Query: 521 TNSLDTANTLTRALRAGTVWVNCFDVFDAAIPFGGYKMSGIGREKGVDSLKNYLQVKAVV 342
T ++D ANT++R++RAG +W+NC+ FD PFGGYKMSG G++ G+D+L YLQ K VV
Sbjct: 435 TKNIDVANTVSRSIRAGAIWINCYFAFDPDAPFGGYKMSGFGKDMGMDALDKYLQTKTVV 494
Query: 341 TPIKNAAWL 315
TP+ N WL
Sbjct: 495 TPLYNTPWL 503
Database: /db/uniprot/tmp/swall
Posted date: Mar 5, 2004 7:45 PM
Number of letters in database: 442,889,342
Number of sequences in database: 1,395,590
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,740,744,100
Number of Sequences: 1395590
Number of extensions: 38037943
Number of successful extensions: 115957
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 100283
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 111174
length of database: 442,889,342
effective HSP length: 131
effective length of database: 260,067,052
effective search space used: 157600633512
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)

